A homozygous FANCM mutation underlies a familial case of non-syndromic primary ovarian insufficiency
- PMID: 29231814
- PMCID: PMC5764568
- DOI: 10.7554/eLife.30490
A homozygous FANCM mutation underlies a familial case of non-syndromic primary ovarian insufficiency
Abstract
Primary Ovarian Insufficiency (POI) affects ~1% of women under forty. Exome sequencing of two Finnish sisters with non-syndromic POI revealed a homozygous mutation in FANCM, leading to a truncated protein (p.Gln1701*). FANCM is a DNA-damage response gene whose heterozygous mutations predispose to breast cancer. Compared to the mother's cells, the patients' lymphocytes displayed higher levels of basal and mitomycin C (MMC)-induced chromosomal abnormalities. Their lymphoblasts were hypersensitive to MMC and MMC-induced monoubiquitination of FANCD2 was impaired. Genetic complementation of patient's cells with wild-type FANCM improved their resistance to MMC re-establishing FANCD2 monoubiquitination. FANCM was more strongly expressed in human fetal germ cells than in somatic cells. FANCM protein was preferentially expressed along the chromosomes in pachytene cells, which undergo meiotic recombination. This mutation may provoke meiotic defects leading to a depleted follicular stock, as in Fancm-/- mice. Our findings document the first Mendelian phenotype due to a biallelic FANCM mutation.
Keywords: FANCM; Mitomycin C; exome sequencing; human; human biology; medicine; mutation; primary ovarian insufficiency; replication inhibitor.
Conflict of interest statement
No competing interests declared.
Figures

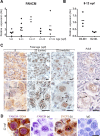
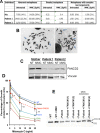
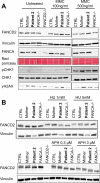
References
-
- Bakker ST, van de Vrugt HJ, Rooimans MA, Oostra AB, Steltenpool J, Delzenne-Goette E, van der Wal A, van der Valk M, Joenje H, te Riele H, de Winter JP. Fancm-deficient mice reveal unique features of Fanconi anemia complementation group M. Human Molecular Genetics. 2009;18:3484–3495. doi: 10.1093/hmg/ddp297. - DOI - PubMed
-
- Bourseguin J, Bonet C, Renaud E, Pandiani C, Boncompagni M, Giuliano S, Pawlikowska P, Karmous-Benailly H, Ballotti R, Rosselli F, Bertolotto C. FANCD2 functions as a critical factor downstream of MiTF to maintain the proliferation and survival of melanoma cells. Scientific Reports. 2016;6:36539. doi: 10.1038/srep36539. - DOI - PMC - PubMed
-
- Catucci I, Osorio A, Arver B, Neidhardt G, Bogliolo M, Zanardi F, Riboni M, Minardi S, Pujol R, Azzollini J, Peissel B, Manoukian S, De Vecchi G, Casola S, Hauke J, Richters L, Rhiem K, Schmutzler RK, Wallander K, Törngren T, Borg Åke, Radice P, Surrallés J, Hahnen E, Ehrencrona H, Kvist A, Benitez J, Peterlongo P. Individuals with FANCM biallelic mutations do not develop Fanconi anemia, but show risk for breast cancer, chemotherapy toxicity and may display chromosome fragility. Genetics in Medicine. 2017;358 doi: 10.1038/gim.2017.123. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

