Maillard Proteomics: Opening New Pages
- PMID: 29231845
- PMCID: PMC5751279
- DOI: 10.3390/ijms18122677
Maillard Proteomics: Opening New Pages
Abstract
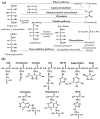
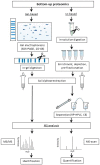
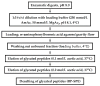
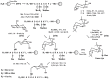
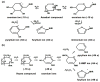
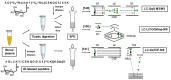
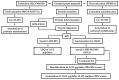
Protein glycation is a ubiquitous non-enzymatic post-translational modification, formed by reaction of protein amino and guanidino groups with carbonyl compounds, presumably reducing sugars and α-dicarbonyls. Resulting advanced glycation end products (AGEs) represent a highly heterogeneous group of compounds, deleterious in mammals due to their pro-inflammatory effect, and impact in pathogenesis of diabetes mellitus, Alzheimer's disease and ageing. The body of information on the mechanisms and pathways of AGE formation, acquired during the last decades, clearly indicates a certain site-specificity of glycation. It makes characterization of individual glycation sites a critical pre-requisite for understanding in vivo mechanisms of AGE formation and developing adequate nutritional and therapeutic approaches to reduce it in humans. In this context, proteomics is the methodology of choice to address site-specific molecular changes related to protein glycation. Therefore, here we summarize the methods of Maillard proteomics, specifically focusing on the techniques providing comprehensive structural and quantitative characterization of glycated proteome. Further, we address the novel break-through areas, recently established in the field of Maillard research, i.e., in vitro models based on synthetic peptides, site-based diagnostics of metabolism-related diseases (e.g., diabetes mellitus), proteomics of anti-glycative defense, and dynamics of plant glycated proteome during ageing and response to environmental stress.
Keywords: advanced glycation end products (AGEs); bottom-up proteomics; glycation; glyoxalase; model synthetic peptides; plant glycation; post-translational modifications; proteomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Amadori M. The product of the condensation of glucose and p-phenetidine. Atti Della R. Accad. Naz. Dei Lincei. 1929;9:68–73.
-
- Heyns K., Noack H. Die Umsetzung von d-Fructose mit l-Lysin and l-Arginin und dered Beziehung zu nichtenzymatischen Bräunungsreaktion. Chem. Ber. 1962;95:720–727. doi: 10.1002/cber.19620950323. - DOI
-
- Hodge J.E. The Amadori rearrangement. Adv. Carbohydr. Chem. 1955;10:169–205. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

