High-Throughput Screening Approach for Identifying Compounds That Inhibit Nonhomologous End Joining
- PMID: 29232168
- PMCID: PMC5997544
- DOI: 10.1177/2472555217746324
High-Throughput Screening Approach for Identifying Compounds That Inhibit Nonhomologous End Joining
Abstract
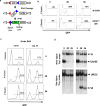
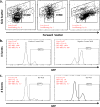
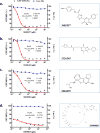
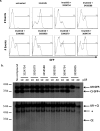
DNA double-strand breaks (DSBs) are repaired primarily by homologous recombination (HR) or nonhomologous end joining (NHEJ). Compounds that modulate HR have shown promise as cancer therapeutics. The V(D)J recombination reaction, which assembles antigen receptor genes in lymphocytes, is initiated by the introduction of DNA DSBs at two recombining gene segments by the RAG endonuclease, followed by the NHEJ-mediated repair of these DSBs. Here, using HyperCyt automated flow cytometry, we develop a robust high-throughput screening (HTS) assay for NHEJ that utilizes engineered pre-B-cell lines where the V(D)J recombination reaction can be induced and monitored at a single-cell level. This approach, novel in processing four 384-well plates at a time in parallel, was used to screen the National Cancer Institute NeXT library to identify compounds that inhibit V(D)J recombination and NHEJ. Assessment of cell light scattering characteristics at the primary HTS stage (83,536 compounds) enabled elimination of 60% of apparent hits as false positives. Although all the active compounds that we identified had an inhibitory effect on RAG cleavage, we have established this as an approach that could identify compounds that inhibit RAG cleavage or NHEJ using new chemical libraries.
Keywords: cancer and cancer drugs; cell-based assays; immune system diseases; oncology.
Figures

References
-
- Bryant HE, Schultz N, Thomas HD, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. - PubMed
-
- Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

