De novo transcriptome assembly from flower buds of dioecious, gynomonoecious and chemically masculinized female Coccinia grandis reveals genes associated with sex expression and modification
- PMID: 29233089
- PMCID: PMC5727884
- DOI: 10.1186/s12870-017-1187-z
De novo transcriptome assembly from flower buds of dioecious, gynomonoecious and chemically masculinized female Coccinia grandis reveals genes associated with sex expression and modification
Abstract
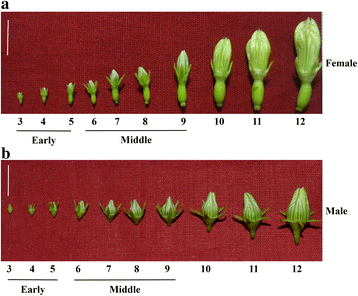
Background: Coccinia grandis (ivy gourd), is a dioecious member of Cucurbitaceae having heteromorphic sex chromosomes. Chromosome constitution of male and female plants of C. grandis is 22A + XY and 22A + XX respectively. Earlier we showed that a unique gynomonoecious form of C. grandis (22A + XX) also exists in nature bearing morphologically hermaphrodite flowers (GyM-H). Additionally, application of silver nitrate (AgNO3) on female plants induces stamen development leading to the formation of morphologically hermaphrodite flowers (Ag-H) despite the absence of Y-chromosome. Due to the unavailability of genome sequence and the slow pace at which sex-linked genes are identified, sex expression and modification in C. grandis are not well understood.
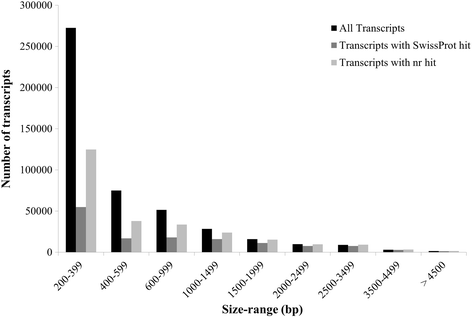
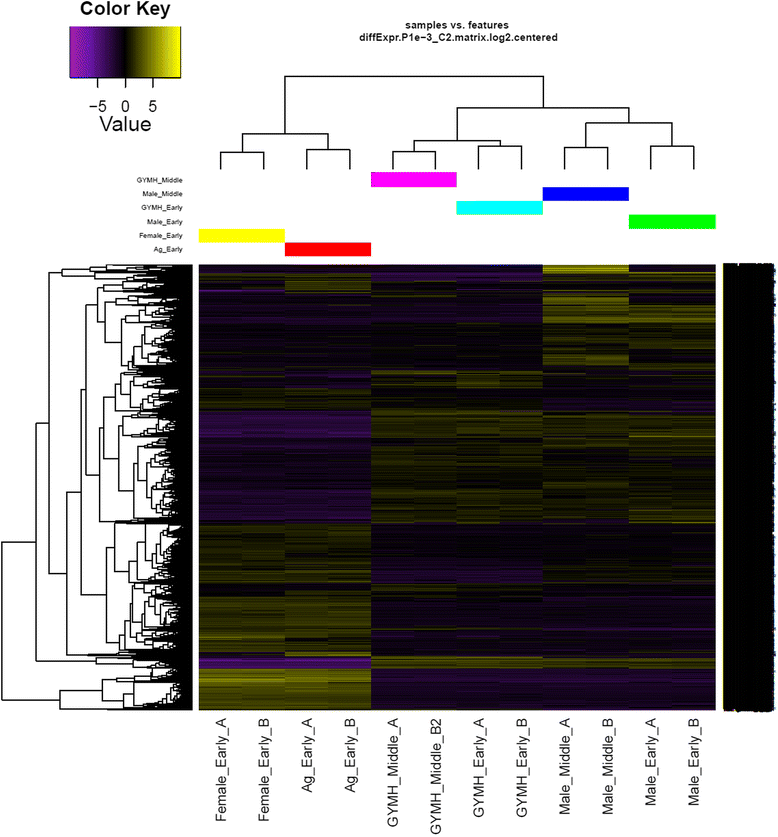
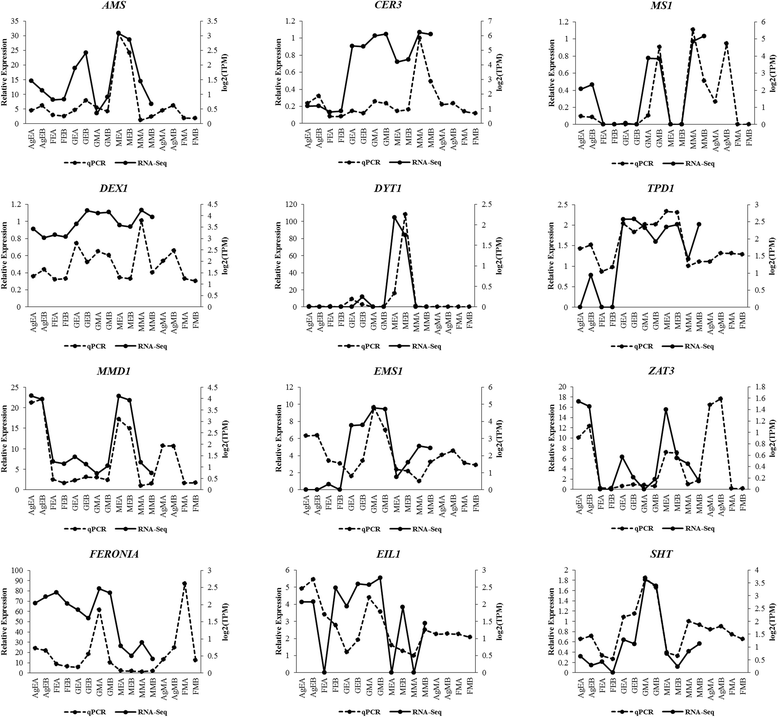
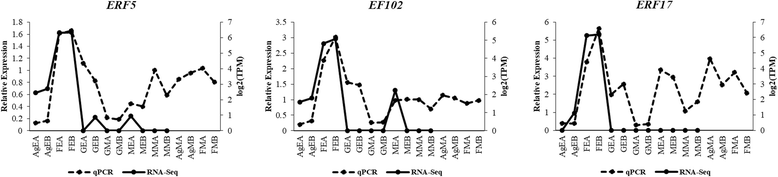
Results: We have carried out a comprehensive RNA-Seq study from early-staged male, female, GyM-H, and Ag-H as well as middle-staged male and GyM-H flower buds. A de novo transcriptome was assembled using Trinity and annotated by BLAST2GO and Trinotate pipelines. The assembled transcriptome consisted of 467,233 'Trinity Transcripts' clustering into 378,860 'Trinity Genes'. Female_Early_vs_Male_Early, Ag_Early_vs_Female_Early, and GyM-H_Middle_vs_Male_Middle comparisons exhibited 35,694, 3574, and 14,954 differentially expressed transcripts respectively. Further, qRT-PCR analysis of selected candidate genes validated digital gene expression profiling results. Interestingly, ethylene response-related genes were found to be upregulated in female buds compared to male buds. Also, we observed that AgNO3 treatment suppressed ethylene responses in Ag-H flowers by downregulation of ethylene-responsive transcription factors leading to stamen development. Further, GO terms related to stamen development were enriched in early-staged male, GyM-H, and Ag-H buds compared to female buds supporting the fact that stamen growth gets arrested in female flowers.
Conclusions: Suppression of ethylene responses in both male and Ag-H compared to female buds suggests a probable role of ethylene in stamen suppression similar to monoecious cucurbits such as melon and cucumber. Also, pollen fertility associated GO terms were depleted in middle-staged GyM-H buds compared to male buds indicating the necessity of Y-chromosome for pollen fertility. Overall, this study would enable identification of new sex-biased genes for further investigation of stamen arrest, pollen fertility, and AgNO3-mediated sex modification.
Keywords: Coccinia grandis; De novo transcriptome; Dioecious; Ethylene; Gynomonoecious; Pollen fertility; Silver nitrate; Stamen arrest.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures




References
-
- Pfent C, Pobursky KJ, Sather DN, Golenberg EM. Characterization of SpAPETALA3 and SpPISTILLATA, B class floral identity genes in Spinacia oleracea, and their relationship to sexual dimorphism. Dev Genes Evol. 2005;215(3):132–42. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

