Selection, characterization, and thermal stabilization of llama single domain antibodies towards Ebola virus glycoprotein
- PMID: 29233140
- PMCID: PMC5726015
- DOI: 10.1186/s12934-017-0837-z
Selection, characterization, and thermal stabilization of llama single domain antibodies towards Ebola virus glycoprotein
Abstract
Background: A key advantage of recombinant antibody technology is the ability to optimize and tailor reagents. Single domain antibodies (sdAbs), the recombinantly produced variable domains derived from camelid and shark heavy chain antibodies, provide advantages of stability and solubility and can be further engineered to enhance their properties. In this study, we generated sdAbs specific for Ebola virus envelope glycoprotein (GP) and increased their stability to expand their utility for use in austere locals. Ebola virus is extremely virulent and causes fatal hemorrhagic fever in ~ 50 percent of the cases. The viral GP binds to host cell receptors to facilitate viral entry and thus plays a critical role in pathogenicity.
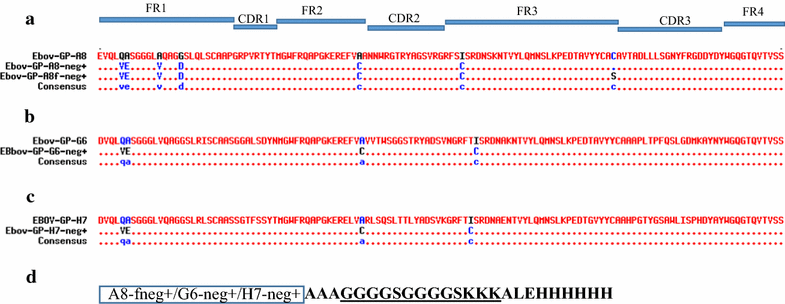
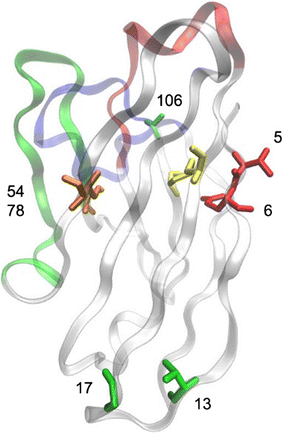
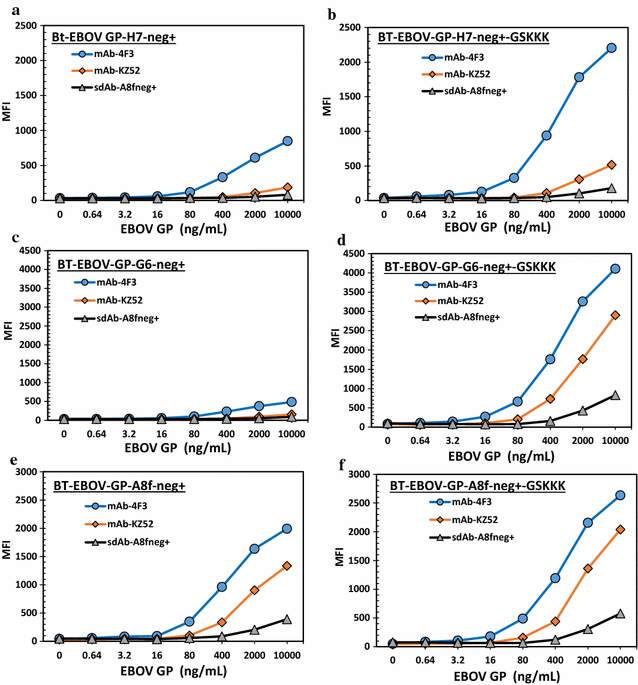
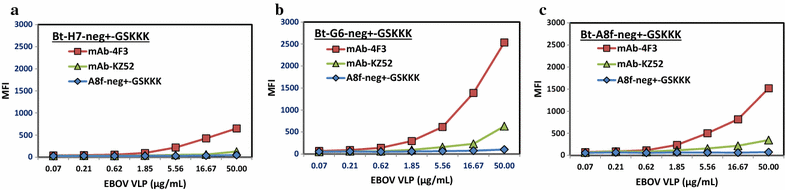
Results: An immune phage display library containing more than 107 unique clones was developed from a llama immunized with a combination of killed Ebola virus and recombinantly produced GP. We panned the library to obtain GP binding sdAbs and isolated sdAbs from 5 distinct sequence families. Three GP binders with dissociation constants ranging from ~ 2 to 20 nM, and melting temperatures from ~ 57 to 72 °C were selected for protein engineering in order to increase their stability through a combination of consensus sequence mutagenesis and the addition of a non-canonical disulfide bond. These changes served to increase the melting temperatures of the sdAbs by 15-17 °C. In addition, fusion of a short positively charged tail to the C-terminus which provided ideal sites for the chemical modification of these sdAbs resulted in improved limits of detection of GP and Ebola virus like particles while serving as tracer antibodies.
Conclusions: SdAbs specific for Ebola GP were selected and their stability and functionality were improved utilizing protein engineering. Thermal stability of antibody reagents may be of particular importance when operating in austere locations that lack reliable refrigeration. Future efforts can evaluate the potential of these isolated sdAbs as candidates for diagnostic or therapeutic applications for Ebola.
Keywords: Antibody engineering; Ebola virus; Glycoprotein; Single domain antibodies; Virus like particles.
Figures

Similar articles
-
Negative tail fusions can improve ruggedness of single domain antibodies.Protein Expr Purif. 2014 Mar;95:226-32. doi: 10.1016/j.pep.2014.01.003. Epub 2014 Jan 15. Protein Expr Purif. 2014. PMID: 24440507
-
Physicochemical improvement of rabbit derived single-domain antibodies by substitutions with amino acids conserved in camelid antibodies.J Biosci Bioeng. 2018 Jun;125(6):654-661. doi: 10.1016/j.jbiosc.2018.01.006. Epub 2018 Feb 15. J Biosci Bioeng. 2018. PMID: 29398547
-
Importance of Hypervariable Region 2 for Stability and Affinity of a Shark Single-Domain Antibody Specific for Ebola Virus Nucleoprotein.PLoS One. 2016 Aug 5;11(8):e0160534. doi: 10.1371/journal.pone.0160534. eCollection 2016. PLoS One. 2016. PMID: 27494523 Free PMC article.
-
Enhancing Stability of Camelid and Shark Single Domain Antibodies: An Overview.Front Immunol. 2017 Jul 25;8:865. doi: 10.3389/fimmu.2017.00865. eCollection 2017. Front Immunol. 2017. PMID: 28791022 Free PMC article. Review.
-
[Research progress on ebola virus glycoprotein].Bing Du Xue Bao. 2013 Mar;29(2):233-7. Bing Du Xue Bao. 2013. PMID: 23757858 Review. Chinese.
Cited by
-
Therapeutic applications of nanobodies against SARS-CoV-2 and other viral infections: Current update.Int J Biol Macromol. 2023 Feb 28;229:70-80. doi: 10.1016/j.ijbiomac.2022.12.284. Epub 2022 Dec 28. Int J Biol Macromol. 2023. PMID: 36586649 Free PMC article. Review.
-
High affinity nanobodies block SARS-CoV-2 spike receptor binding domain interaction with human angiotensin converting enzyme.Sci Rep. 2020 Dec 22;10(1):22370. doi: 10.1038/s41598-020-79036-0. Sci Rep. 2020. PMID: 33353972 Free PMC article.
-
A field effect transistor modified with reduced graphene oxide for immunodetection of Ebola virus.Mikrochim Acta. 2019 Mar 7;186(4):223. doi: 10.1007/s00604-019-3256-5. Mikrochim Acta. 2019. PMID: 30847625
-
Nanobodies in the fight against infectious diseases: repurposing nature's tiny weapons.World J Microbiol Biotechnol. 2024 May 21;40(7):209. doi: 10.1007/s11274-024-03990-4. World J Microbiol Biotechnol. 2024. PMID: 38771414 Free PMC article. Review.
-
Prospects of Neutralizing Nanobodies Against SARS-CoV-2.Front Immunol. 2021 May 28;12:690742. doi: 10.3389/fimmu.2021.690742. eCollection 2021. Front Immunol. 2021. PMID: 34122456 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

