Molecular Anatomy of the Developing Human Retina
- PMID: 29233477
- PMCID: PMC5776731
- DOI: 10.1016/j.devcel.2017.10.029
Molecular Anatomy of the Developing Human Retina
Abstract
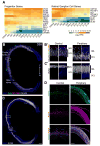
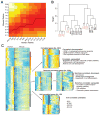
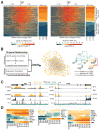
Clinical and genetic heterogeneity associated with retinal diseases makes stem-cell-based therapies an attractive strategy for personalized medicine. However, we have limited understanding of the timing of key events in the developing human retina, and in particular the factors critical for generating the unique architecture of the fovea and surrounding macula. Here we define three key epochs in the transcriptome dynamics of human retina from fetal day (D) 52 to 136. Coincident histological analyses confirmed the cellular basis of transcriptional changes and highlighted the dramatic acceleration of development in the fovea compared with peripheral retina. Human and mouse retinal transcriptomes show remarkable similarity in developmental stages, although morphogenesis was greatly expanded in humans. Integration of DNA accessibility data allowed us to reconstruct transcriptional networks controlling photoreceptor differentiation. Our studies provide insights into human retinal development and serve as a resource for molecular staging of human stem-cell-derived retinal organoids.
Keywords: development; fetal; fovea; gene network; human; macula; organoids; photoreceptor; retina; transcriptome.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures





References
-
- Bumsted O’Brien KM, Cheng H, Jiang Y, Schulte D, Swaroop A, Hendrickson AE. Expression of photoreceptor-specific nuclear receptor NR2E3 in rod photoreceptors of fetal human retina. Investigative ophthalmology & visual science. 2004;45:2807–2812. - PubMed
-
- Cayouette M, Poggi L, Harris WA. Lineage in the vertebrate retina. Trends Neurosci. 2006;29:563–570. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

