Breast Cancers Activate Stromal Fibroblast-Induced Suppression of Progenitors in Adjacent Normal Tissue
- PMID: 29233553
- PMCID: PMC5768884
- DOI: 10.1016/j.stemcr.2017.11.002
Breast Cancers Activate Stromal Fibroblast-Induced Suppression of Progenitors in Adjacent Normal Tissue
Abstract
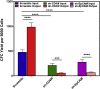
Human breast cancer cells are known to activate adjacent "normal-like" cells to enhance their own growth, but the cellular and molecular mechanisms involved are poorly understood. We now show by both phenotypic and functional measurements that normal human mammary progenitor cells are significantly under-represented in the mammary epithelium of patients' tumor-adjacent tissue (TAT). Interestingly, fibroblasts isolated from TAT samples showed a reduced ability to support normal EGF-stimulated mammary progenitor cell proliferation in vitro via their increased secretion of transforming growth factor β. In contrast, TAT fibroblasts promoted the proliferation of human breast cancer cells when these were co-transplanted in immunodeficient mice. The discovery of a common stromal cell-mediated mechanism that has opposing growth-suppressive and promoting effects on normal and malignant human breast cells and also extends well beyond currently examined surgical margins has important implications for disease recurrence and its prevention.
Keywords: TGF-β; activated stromal fibroblasts; breast cancer; suppression of normal breast tissue progenitors; tumor-adjacent tissue.
Copyright © 2017 The Authors. Published by Elsevier Inc. All rights reserved.
Figures







Similar articles
-
Fibroblasts direct differentiation of human breast epithelial progenitors.Breast Cancer Res. 2020 Sep 29;22(1):102. doi: 10.1186/s13058-020-01344-0. Breast Cancer Res. 2020. PMID: 32993755 Free PMC article.
-
JNK1 stress signaling is hyper-activated in high breast density and the tumor stroma: connecting fibrosis, inflammation, and stemness for cancer prevention.Cell Cycle. 2014;13(4):580-99. doi: 10.4161/cc.27379. Epub 2013 Dec 5. Cell Cycle. 2014. PMID: 24434780 Free PMC article.
-
Elevated CXCL1 expression in breast cancer stroma predicts poor prognosis and is inversely associated with expression of TGF-β signaling proteins.BMC Cancer. 2014 Oct 24;14:781. doi: 10.1186/1471-2407-14-781. BMC Cancer. 2014. PMID: 25344051 Free PMC article.
-
Early and late events in the development of human breast cancer.Basic Life Sci. 1991;57:329-37; discussion 337-40. doi: 10.1007/978-1-4684-5994-4_27. Basic Life Sci. 1991. PMID: 1814293 Review.
-
Breast tumor and stromal cell responses to TGF-β and hypoxia in matrix deposition.Matrix Biol. 2013 Mar 11;32(2):95-105. doi: 10.1016/j.matbio.2012.11.016. Epub 2012 Dec 20. Matrix Biol. 2013. PMID: 23262216 Free PMC article. Review.
Cited by
-
Role of the Microenvironment in Regulating Normal and Cancer Stem Cell Activity: Implications for Breast Cancer Progression and Therapy Response.Cancers (Basel). 2019 Aug 24;11(9):1240. doi: 10.3390/cancers11091240. Cancers (Basel). 2019. PMID: 31450577 Free PMC article. Review.
-
Endometrial Cancer-Adjacent Tissues Express Higher Levels of Cancer-Promoting Genes than the Matched Tumors.Genes (Basel). 2022 Sep 8;13(9):1611. doi: 10.3390/genes13091611. Genes (Basel). 2022. PMID: 36140779 Free PMC article.
-
Instant fluorescence lifetime imaging microscopy reveals mechano-metabolic reprogramming of stromal cells in breast cancer peritumoral microenvironments.bioRxiv [Preprint]. 2025 May 30:2025.05.28.656717. doi: 10.1101/2025.05.28.656717. bioRxiv. 2025. PMID: 40501613 Free PMC article. Preprint.
-
Residual Risk of Ipsilateral Tumor Recurrence in Patients Who Achieved Clear Lumpectomy Margins After Repeated Resection.J Breast Cancer. 2023 Dec;26(6):558-571. doi: 10.4048/jbc.2023.26.e46. Epub 2023 Oct 17. J Breast Cancer. 2023. PMID: 37985383 Free PMC article.
-
The interplay of quaternary ammonium lipid structure and protein corona on lung-specific mRNA delivery by selective organ targeting (SORT) nanoparticles.J Control Release. 2023 Sep;361:361-372. doi: 10.1016/j.jconrel.2023.07.058. Epub 2023 Aug 10. J Control Release. 2023. PMID: 37536547 Free PMC article.
References
-
- Allinen M., Beroukhim R., Cai L., Brennan C., Lahti-Domenici J., Huang H., Porter D., Hu M., Chin L., Richardson A. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004;6:17–32. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

