Multiplexed in vivo homology-directed repair and tumor barcoding enables parallel quantification of Kras variant oncogenicity
- PMID: 29233960
- PMCID: PMC5727199
- DOI: 10.1038/s41467-017-01519-y
Multiplexed in vivo homology-directed repair and tumor barcoding enables parallel quantification of Kras variant oncogenicity
Abstract
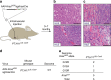
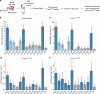
Large-scale genomic analyses of human cancers have cataloged somatic point mutations thought to initiate tumor development and sustain cancer growth. However, determining the functional significance of specific alterations remains a major bottleneck in our understanding of the genetic determinants of cancer. Here, we present a platform that integrates multiplexed AAV/Cas9-mediated homology-directed repair (HDR) with DNA barcoding and high-throughput sequencing to simultaneously investigate multiple genomic alterations in de novo cancers in mice. Using this approach, we introduce a barcoded library of non-synonymous mutations into hotspot codons 12 and 13 of Kras in adult somatic cells to initiate tumors in the lung, pancreas, and muscle. High-throughput sequencing of barcoded Kras HDR alleles from bulk lung and pancreas reveals surprising diversity in Kras variant oncogenicity. Rapid, cost-effective, and quantitative approaches to simultaneously investigate the function of precise genomic alterations in vivo will help uncover novel biological and clinically actionable insights into carcinogenesis.
Conflict of interest statement
Stanford University has filed a patent (U.S. Provisional Application 62/481,067) related to this work in which I.P.W., C.D.M., D.A.P., and M.M.W. are listed as co-inventors. The remaining authors declare no competing financial interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

