PEPD is a pivotal regulator of p53 tumor suppressor
- PMID: 29233996
- PMCID: PMC5727116
- DOI: 10.1038/s41467-017-02097-9
PEPD is a pivotal regulator of p53 tumor suppressor
Abstract
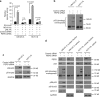
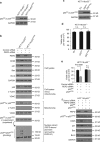
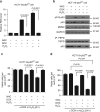
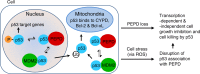
p53 tumor suppressor responds to various cellular stresses and regulates cell fate. Here, we show that peptidase D (PEPD) binds and suppresses over half of nuclear and cytoplasmic p53 under normal conditions, independent of its enzymatic activity. Eliminating PEPD causes cell death and tumor regression due to p53 activation. PEPD binds to the proline-rich domain in p53, which inhibits phosphorylation of nuclear p53 and MDM2-mediated mitochondrial translocation of nuclear and cytoplasmic p53. However, the PEPD-p53 complex is critical for p53 response to stress, as stress signals doxorubicin and H2O2 each must free p53 from PEPD in order to achieve robust p53 activation, which is mediated by reactive oxygen species. Thus, PEPD stores p53 for the stress response, but this also renders cells dependent on PEPD for survival, as it suppresses p53. This finding provides further understanding of p53 regulation and may have significant implications for the treatment of cancer and other diseases.
Conflict of interest statement
The authors declare no competing financial interests.
Figures






Similar articles
-
Loss of peptidase D binding restores the tumor suppressor functions of oncogenic p53 mutants.Commun Biol. 2021 Dec 8;4(1):1373. doi: 10.1038/s42003-021-02880-x. Commun Biol. 2021. PMID: 34880421 Free PMC article.
-
SPARC functions as an anti-stress factor by inactivating p53 through Akt-mediated MDM2 phosphorylation to promote melanoma cell survival.Oncogene. 2011 Dec 8;30(49):4887-900. doi: 10.1038/onc.2011.198. Epub 2011 Jun 20. Oncogene. 2011. PMID: 21685937
-
Stabilization and activation of p53 induced by Cdk5 contributes to neuronal cell death.J Cell Sci. 2007 Jul 1;120(Pt 13):2259-71. doi: 10.1242/jcs.03468. J Cell Sci. 2007. PMID: 17591690
-
The regulation of MDM2 by multisite phosphorylation--opportunities for molecular-based intervention to target tumours?Semin Cancer Biol. 2010 Feb;20(1):19-28. doi: 10.1016/j.semcancer.2009.10.005. Epub 2009 Nov 6. Semin Cancer Biol. 2010. PMID: 19897041 Review.
-
Cytoplasmic functions of the tumour suppressor p53.Nature. 2009 Apr 30;458(7242):1127-30. doi: 10.1038/nature07986. Nature. 2009. PMID: 19407794 Free PMC article. Review.
Cited by
-
A recombinant human protein targeting HER2 overcomes drug resistance in HER2-positive breast cancer.Sci Transl Med. 2019 Jan 23;11(476):eaav1620. doi: 10.1126/scitranslmed.aav1620. Sci Transl Med. 2019. PMID: 30674653 Free PMC article.
-
Extracellular Prolidase (PEPD) Induces Anabolic Processes through EGFR, β1-integrin, and IGF-1R Signaling Pathways in an Experimental Model of Wounded Fibroblasts.Int J Mol Sci. 2021 Jan 19;22(2):942. doi: 10.3390/ijms22020942. Int J Mol Sci. 2021. PMID: 33477899 Free PMC article.
-
Circadian clock protein CRY1 prevents paclitaxel‑induced senescence of bladder cancer cells by promoting p53 degradation.Oncol Rep. 2021 Mar;45(3):1033-1043. doi: 10.3892/or.2020.7914. Epub 2020 Dec 30. Oncol Rep. 2021. PMID: 33650658 Free PMC article.
-
Prolidase-proline oxidase axis is engaged in apoptosis induction by birch buds flavonol santin in endometrial adenocarcinoma cell line.Front Mol Biosci. 2023 Sep 6;10:1247536. doi: 10.3389/fmolb.2023.1247536. eCollection 2023. Front Mol Biosci. 2023. PMID: 37745688 Free PMC article.
-
Targeting p53 pathways: mechanisms, structures, and advances in therapy.Signal Transduct Target Ther. 2023 Mar 1;8(1):92. doi: 10.1038/s41392-023-01347-1. Signal Transduct Target Ther. 2023. PMID: 36859359 Free PMC article.
References
-
- Bergmann M, Fruton JS. On proteolytic enzymes: XII. Regarding the specificity of aminopeptidase and carboxypeptidase. A new type of enzyme in the intestinal tract. J. Biol. Chem. 1937;117:189–202.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

