Rokubacteria: Genomic Giants among the Uncultured Bacterial Phyla
- PMID: 29234309
- PMCID: PMC5712423
- DOI: 10.3389/fmicb.2017.02264
Rokubacteria: Genomic Giants among the Uncultured Bacterial Phyla
Abstract
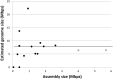
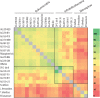
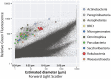
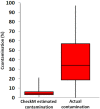
Recent advances in single-cell genomic and metagenomic techniques have facilitated the discovery of numerous previously unknown, deep branches of the tree of life that lack cultured representatives. Many of these candidate phyla are composed of microorganisms with minimalistic, streamlined genomes lacking some core metabolic pathways, which may contribute to their resistance to growth in pure culture. Here we analyzed single-cell genomes and metagenome bins to show that the "Candidate phylum Rokubacteria," formerly known as SPAM, represents an interesting exception, by having large genomes (6-8 Mbps), high GC content (66-71%), and the potential for a versatile, mixotrophic metabolism. We also observed an unusually high genomic heterogeneity among individual Rokubacteria cells in the studied samples. These features may have contributed to the limited recovery of sequences of this candidate phylum in prior cultivation and metagenomic studies. Our analyses suggest that Rokubacteria are distributed globally in diverse terrestrial ecosystems, including soils, the rhizosphere, volcanic mud, oil wells, aquifers, and the deep subsurface, with no reports from marine environments to date.
Keywords: microbial dark matter; microbial ecology; microbial evolution; microbial genomics; uncultivated bacteria.
Figures


References
-
- Becraft E. D., Dodsworth J. A., Murugapiran S. K., Ohlsson J. I., Briggs B. R., Kanbar J., et al. . (2015). Single-cell-genomics-facilitated read binning of candidate phylum EM19 genomes from geothermal spring metagenomes. Appl. Environ. Microbiol. 82, 992–1003. 10.1128/AEM.03140-15 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

