Bacterial flagellar axial structure and its construction
- PMID: 29235079
- PMCID: PMC5899737
- DOI: 10.1007/s12551-017-0378-z
Bacterial flagellar axial structure and its construction
Abstract
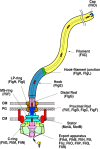
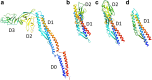
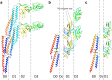
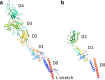
The bacterial flagellum is a motile organelle composed of thousands of protein subunits. The filamentous part that extends from the cell membrane is called the axial structure and consists of three major parts, the filament, hook, and rod, and other minor components. Each of the three main parts shares a similar self-assembly mechanism and a common basic architecture of subunit arrangement while showing quite distinct mechanical properties to achieve its specific function. Structural and molecular mechanisms to produce these various mechanical properties of the axial structure, such as the filament, the hook, and the rod, have been revealed by the complementary use of X-ray crystallography and cryo-electron microscopy. In addition, the mechanism of growth of the axial structure is beginning to be revealed based on the molecular structure.
Keywords: Axial structure; Bacterial flagellum; Cryo-electron microscopy; X-ray crystallography.
Conflict of interest statement
Conflict of interest
Katsumi Imada declares that he has no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by the author.
Figures



References
-
- Asakura S. Polymerization of flagellin and polymorphism of flagella. Advan Biophys (Japan) 1970;1:99–155. - PubMed
-
- Buist G, Kok J, Leenhouts KJ, Dabrowska M, Venema G, Haandrikman AJ. Molecular cloning and nucleotide sequence of the gene encoding the major peptidoglycan hydrolase of Lactococcus lactis, a muramidase needed for cell separation. J Bacteriol. 1995;177:1554–1563. doi: 10.1128/jb.177.6.1554-1563.1995. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

