Inactivation of the dnaK gene in Clostridium difficile 630 Δerm yields a temperature-sensitive phenotype and increases biofilm-forming ability
- PMID: 29235503
- PMCID: PMC5727486
- DOI: 10.1038/s41598-017-17583-9
Inactivation of the dnaK gene in Clostridium difficile 630 Δerm yields a temperature-sensitive phenotype and increases biofilm-forming ability
Abstract
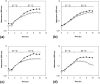
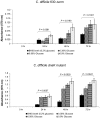
Clostridium difficile infection is a growing problem in healthcare settings worldwide and results in a considerable socioeconomic impact. New hypervirulent strains and acquisition of antibiotic resistance exacerbates pathogenesis; however, the survival strategy of C. difficile in the challenging gut environment still remains incompletely understood. We previously reported that clinically relevant heat-stress (37-41 °C) resulted in a classical heat-stress response with up-regulation of cellular chaperones. We used ClosTron to construct an insertional mutation in the dnaK gene of C. difficile 630 Δerm. The dnaK mutant exhibited temperature sensitivity, grew more slowly than C. difficile 630 Δerm and was less thermotolerant. Furthermore, the mutant was non-motile, had 4-fold lower expression of the fliC gene and lacked flagella on the cell surface. Mutant cells were some 50% longer than parental strain cells, and at optimal growth temperatures, they exhibited a 4-fold increase in the expression of class I chaperone genes including GroEL and GroES. Increased chaperone expression, in addition to the non-flagellated phenotype of the mutant, may account for the increased biofilm formation observed. Overall, the phenotype resulting from dnaK disruption is more akin to that observed in Escherichia coli dnaK mutants, rather than those in the Gram-positive model organism Bacillus subtilis.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Similar articles
-
Important role of class I heat shock genes hrcA and dnaK in the heat shock response and the response to pH and NaCl stress of group I Clostridium botulinum strain ATCC 3502.Appl Environ Microbiol. 2011 May;77(9):2823-30. doi: 10.1128/AEM.02633-10. Epub 2011 Mar 4. Appl Environ Microbiol. 2011. PMID: 21378058 Free PMC article.
-
Recognizability of heterologous co-chaperones with Streptococcus intermedius DnaK and Escherichia coli DnaK.Microbiol Immunol. 2018 Nov;62(11):681-693. doi: 10.1111/1348-0421.12651. Epub 2018 Nov 5. Microbiol Immunol. 2018. PMID: 30239035
-
Role of Streptococcus intermedius DnaK chaperone system in stress tolerance and pathogenicity.Cell Stress Chaperones. 2012 Jan;17(1):41-55. doi: 10.1007/s12192-011-0284-4. Epub 2011 Aug 6. Cell Stress Chaperones. 2012. PMID: 21822788 Free PMC article.
-
Biofilm regulation in Clostridioides difficile: Novel systems linked to hypervirulence.PLoS Pathog. 2021 Sep 9;17(9):e1009817. doi: 10.1371/journal.ppat.1009817. eCollection 2021 Sep. PLoS Pathog. 2021. PMID: 34499698 Free PMC article. Review.
-
Type IV pili are involved in phenotypes associated with Clostridioides difficile pathogenesis.Crit Rev Microbiol. 2024 Nov;50(6):1011-1019. doi: 10.1080/1040841X.2023.2235002. Epub 2023 Jul 15. Crit Rev Microbiol. 2024. PMID: 37452617 Review.
Cited by
-
Clostridioides difficile biofilms: A mechanism of persistence in the gut?PLoS Pathog. 2021 Mar 11;17(3):e1009348. doi: 10.1371/journal.ppat.1009348. eCollection 2021 Mar. PLoS Pathog. 2021. PMID: 33705497 Free PMC article. Review. No abstract available.
-
Antibiotic Resistance and Biofilm Production Capacity in Clostridioides difficile.Front Cell Infect Microbiol. 2021 Aug 4;11:683464. doi: 10.3389/fcimb.2021.683464. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34422678 Free PMC article.
-
The DnaJK chaperone of Bacillus subtilis post-transcriptionally regulates gene expression through the YlxR(RnpM)/RNase P complex.mBio. 2025 Mar 12;16(3):e0405324. doi: 10.1128/mbio.04053-24. Epub 2025 Feb 11. mBio. 2025. PMID: 39932325 Free PMC article.
-
Protein Homeostasis Impairment Alters Phenotypic Heterogeneity of Biofilm Communities.Mol Microbiol. 2025 Jul;124(1):1-19. doi: 10.1111/mmi.15366. Epub 2025 Apr 17. Mol Microbiol. 2025. PMID: 40243034 Free PMC article.
-
Utilizing a reductionist model to study host-microbe interactions in intestinal inflammation.Microbiome. 2021 Nov 3;9(1):215. doi: 10.1186/s40168-021-01161-3. Microbiome. 2021. PMID: 34732258 Free PMC article.
References
-
- Freeman J, et al. Pan-European longitudinal surveillance of antibiotic resistance among prevalent Clostridium difficile ribotypes’ study group. Pan-European longitudinal surveillance of antibiotic resistance among prevalent Clostridium difficile ribotypes. Clin. Microbiol. Infect. 2015;21:248.e9–248.e16. doi: 10.1016/j.cmi.2014.09.017. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

