Convergent evolution of bilaterian nerve cords
- PMID: 29236686
- PMCID: PMC5756474
- DOI: 10.1038/nature25030
Convergent evolution of bilaterian nerve cords
Abstract
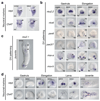
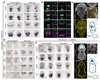
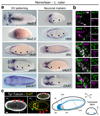
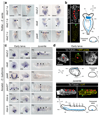
It has been hypothesized that a condensed nervous system with a medial ventral nerve cord is an ancestral character of Bilateria. The presence of similar dorsoventral molecular patterns along the nerve cords of vertebrates, flies, and an annelid has been interpreted as support for this scenario. Whether these similarities are generally found across the diversity of bilaterian neuroanatomies is unclear, and thus the evolutionary history of the nervous system is still contentious. Here we study representatives of Xenacoelomorpha, Rotifera, Nemertea, Brachiopoda, and Annelida to assess the conservation of the dorsoventral nerve cord patterning. None of the studied species show a conserved dorsoventral molecular regionalization of their nerve cords, not even the annelid Owenia fusiformis, whose trunk neuroanatomy parallels that of vertebrates and flies. Our findings restrict the use of molecular patterns to explain nervous system evolution, and suggest that the similarities in dorsoventral patterning and trunk neuroanatomies evolved independently in Bilateria.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

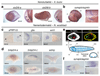


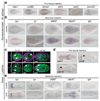
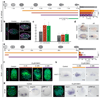

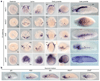



Comment in
-
More than one way to a central nervous system.Nature. 2018 Jan 4;553(7686):34-36. doi: 10.1038/d41586-017-08195-4. Nature. 2018. PMID: 29300031 No abstract available.
-
Animal Evolution: Convergent Nerve Cords?Curr Biol. 2018 Mar 5;28(5):R225-R227. doi: 10.1016/j.cub.2018.01.056. Curr Biol. 2018. PMID: 29510113
References
-
- Schmidt-Rhaesa A, Harzsch S, Purschke G. Structure & Evolution of Invertebrate Nervous Systems. Oxford University Press; 2016.
-
- Arendt D, Tosches MA, Marlow H. From nerve net to nerve ring, nerve cord and brain--evolution of the nervous system. Nat Rev Neurosci. 2016;17:61–72. - PubMed
-
- Hejnol A, Pang K. Xenacoelomorpha's significance for understanding bilaterian evolution. Curr Opin Genet Dev. 2016;39:48–54. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

