Redox-Sensing Under Hypochlorite Stress and Infection Conditions by the Rrf2-Family Repressor HypR in Staphylococcus aureus
- PMID: 29237286
- PMCID: PMC6067689
- DOI: 10.1089/ars.2017.7354
Redox-Sensing Under Hypochlorite Stress and Infection Conditions by the Rrf2-Family Repressor HypR in Staphylococcus aureus
Abstract
Aims: Staphylococcus aureus is a major human pathogen and has to cope with reactive oxygen and chlorine species (ROS, RCS) during infections, which requires efficient protection mechanisms to avoid destruction. Here, we have investigated the changes in the RNA-seq transcriptome by the strong oxidant sodium hypochlorite (NaOCl) in S. aureus USA300 to identify novel redox-sensing mechanisms that provide protection under infection conditions.
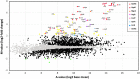
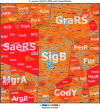
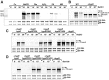
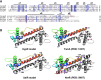
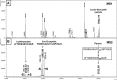
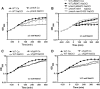
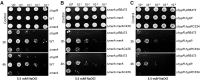
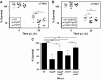
Results: NaOCl stress caused an oxidative stress response in S. aureus as indicated by the induction of the PerR, QsrR, HrcA, and SigmaB regulons in the RNA-seq transcriptome. The hypR-merA (USA300HOU_0588-87) operon was most strongly upregulated under NaOCl stress, which encodes for the Rrf2-family regulator HypR and the pyridine nucleotide disulfide reductase MerA. We have characterized HypR as a novel redox-sensitive repressor that controls MerA expression and directly senses and responds to NaOCl and diamide stress via a thiol-based mechanism in S. aureus. Mutational analysis identified Cys33 and the conserved Cys99 as essential for NaOCl sensing, while Cys99 is also important for repressor activity of HypR in vivo. The redox-sensing mechanism of HypR involves Cys33-Cys99 intersubunit disulfide formation by NaOCl stress both in vitro and in vivo. Moreover, the HypR-controlled flavin disulfide reductase MerA was shown to protect S. aureus against NaOCl stress and increased survival in J774A.1 macrophage infection assays. Conclusion and Innovation: Here, we identified a new member of the widespread Rrf2 family as redox sensor of NaOCl stress in S. aureus that uses a thiol/disulfide switch to regulate defense mechanisms against the oxidative burst under infections in S. aureus. Antioxid. Redox Signal. 29, 615-636.
Keywords: Rrf2; Staphylococcus aureus; hypochlorite stress; redox-sensing regulator.
Conflict of interest statement
No competing financial interests exist.
Figures





Similar articles
-
Staphylococcus aureus responds to allicin by global S-thioallylation - Role of the Brx/BSH/YpdA pathway and the disulfide reductase MerA to overcome allicin stress.Free Radic Biol Med. 2019 Aug 1;139:55-69. doi: 10.1016/j.freeradbiomed.2019.05.018. Epub 2019 May 20. Free Radic Biol Med. 2019. PMID: 31121222
-
Structural insights into the redox-switch mechanism of the MarR/DUF24-type regulator HypR.Nucleic Acids Res. 2012 May;40(9):4178-92. doi: 10.1093/nar/gkr1316. Epub 2012 Jan 11. Nucleic Acids Res. 2012. PMID: 22238377 Free PMC article.
-
Protein S-Bacillithiolation Functions in Thiol Protection and Redox Regulation of the Glyceraldehyde-3-Phosphate Dehydrogenase Gap in Staphylococcus aureus Under Hypochlorite Stress.Antioxid Redox Signal. 2018 Feb 20;28(6):410-430. doi: 10.1089/ars.2016.6897. Epub 2017 Jan 18. Antioxid Redox Signal. 2018. PMID: 27967218 Free PMC article.
-
Thiol-based redox switches in the major pathogen Staphylococcus aureus.Biol Chem. 2020 Nov 23;402(3):333-361. doi: 10.1515/hsz-2020-0272. Print 2021 Feb 23. Biol Chem. 2020. PMID: 33544504 Review.
-
Thiol-based redox switches in prokaryotes.Biol Chem. 2015 May;396(5):415-44. doi: 10.1515/hsz-2015-0102. Biol Chem. 2015. PMID: 25720121 Free PMC article. Review.
Cited by
-
The Catalase KatA Contributes to Microaerophilic H2O2 Priming to Acquire an Improved Oxidative Stress Resistance in Staphylococcus aureus.Antioxidants (Basel). 2022 Sep 12;11(9):1793. doi: 10.3390/antiox11091793. Antioxidants (Basel). 2022. PMID: 36139867 Free PMC article.
-
TrmB Family Transcription Factor as a Thiol-Based Regulator of Oxidative Stress Response.mBio. 2022 Aug 30;13(4):e0063322. doi: 10.1128/mbio.00633-22. Epub 2022 Jul 20. mBio. 2022. PMID: 35856564 Free PMC article.
-
Escherichia coli RclA is a highly active hypothiocyanite reductase.Proc Natl Acad Sci U S A. 2022 Jul 26;119(30):e2119368119. doi: 10.1073/pnas.2119368119. Epub 2022 Jul 22. Proc Natl Acad Sci U S A. 2022. PMID: 35867824 Free PMC article.
-
The MarR-Type Repressor MhqR Confers Quinone and Antimicrobial Resistance in Staphylococcus aureus.Antioxid Redox Signal. 2019 Dec 1;31(16):1235-1252. doi: 10.1089/ars.2019.7750. Epub 2019 Aug 9. Antioxid Redox Signal. 2019. PMID: 31310152 Free PMC article.
-
The two-Cys-type TetR repressor GbaA confers resistance under disulfide and electrophile stress in Staphylococcus aureus.Free Radic Biol Med. 2021 Dec;177:120-131. doi: 10.1016/j.freeradbiomed.2021.10.024. Epub 2021 Oct 19. Free Radic Biol Med. 2021. PMID: 34678418 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
