Determining the target protein localization in 3D using the combination of FIB-SEM and APEX2
- PMID: 29238746
- PMCID: PMC5719812
- DOI: 10.1007/s41048-017-0043-x
Determining the target protein localization in 3D using the combination of FIB-SEM and APEX2
Abstract
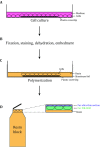
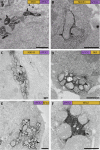
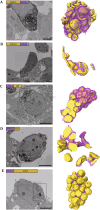
Determining the cellular localization of proteins of interest at nanometer resolution is necessary for elucidating their functions. Besides super-resolution fluorescence microscopy, conventional electron microscopy (EM) combined with immunolabeling or clonable EM tags provides a unique approach to correlate protein localization information and cellular ultrastructural information. However, there are still rare cases of such correlation in three-dimensional (3D) spaces. Here, we developed an approach by combining the focus ion beam scanning electron microscopy (FIB-SEM) and a promising clonable EM tag APEX2 (an enhanced ascorbate peroxidase 2) to determine the target protein localization within 3D cellular ultrastructural context. We further utilized this approach to study the 3D localization of mitochondrial dynamics-related proteins (MiD49/51, Mff, Fis1, and Mfn2) in the cells where the target proteins were overexpressed. We found that all the target proteins were located at the surface of the mitochondrial outer membrane accompanying with mitochondrial clusters. Mid49/51, Mff, and hFis1 spread widely around the mitochondrial surface while Mfn2 only exists at the contact sites.
Keywords: Enhanced ascorbate peroxidase; Focus ion beam scanning electron microscopy; Mitochondrial dynamics; Protein location; Three-dimensional space.
Conflict of interest statement
Compliance with Ethical StandardsYang Shi, Li Wang, Jianguo Zhang, Yujia Zhai, and Fei Sun declare that they have no conflict of interest.This article does not contain any studies with human or animal subjects performed by any of the authors.
Figures
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
