Five key lncRNAs considered as prognostic targets for predicting pancreatic ductal adenocarcinoma
- PMID: 29239017
- PMCID: PMC5947154
- DOI: 10.1002/jcb.26598
Five key lncRNAs considered as prognostic targets for predicting pancreatic ductal adenocarcinoma
Abstract
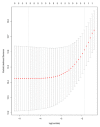
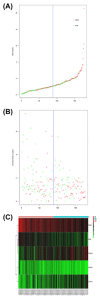
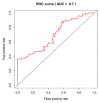
Pancreatic ductal adenocarcinoma (PDAC) has a poor prognosis, and the 5-year survival rate was only 7.7%. To improve prognosis, a screening biomarker for early diagnosis of pancreatic cancer is in urgent need. Long non-coding RNA (lncRNA) expression profiles as potential cancer prognostic biomarkers play critical roles in development of tumorigenesis and metastasis of cancer. However, lncRNA signatures in predicting the survival of a patient with PDAC remain unknown. In the current study, we try to identify potential lncRNA biomarkers and their prognostic values in PDAC. LncRNAs expression profiles and corresponding clinical information for 182 cases with PDAC were acquired from The Cancer Genome Atlas (TCGA). A total of 14 470 lncRNA were identified in the cohort, and 175 PDAC patients had clinical variables. We obtained 108 differential expressed lncRNA via R packages. Univariate and multivariate Cox proportional hazards regression, lasso regression was performed to screen the potential prognostic lncRNA. Five lncRNAs have been recognized to significantly correlate with OS. We established a linear prognostic model of five lncRNA (C9orf139, MIR600HG, RP5-965G21.4, RP11-436K8.1, and CTC-327F10.4) and divided patients into high- and low-risk group according to the prognostic index. The five lncRNAs played independent prognostic biomarkers of OS of PDAC patients and the AUC of the ROC curve for the five lncRNAs signatures prediction 5-year survival was 0.742. In addition, targeted genes of MIR600HG, C9orf139, and CTC-327F10.4 were explored and functional enrichment was also conducted. These results suggested that this five-lncRNAs signature could act as potential prognostic biomarkers in the prediction of PDAC patient's survival.
Keywords: lncRNAs; pancreatic ductal adenocarcinoma; weighted gene co-expression network analysis.
© 2017 The Authors. Journal of Cellular Biochemistry Published by Wiley Periodicals, Inc.
Figures








References
-
- Vogelzang NJ, Benowitz SI, Adams S, et al. Clinical cancer advances 2011: Annual Report on Progress Against Cancer from the American Society of Clinical Oncology. J Clin Oncol. 2012; 30:88–109. - PubMed
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017; 67:7–30. - PubMed
-
- Gloss BS, Dinger ME. The specificity of long noncoding RNA expression. Biochim Biophys Acta. 2016; 1859:16–22. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

