Protein depalmitoylases
- PMID: 29239216
- PMCID: PMC6009847
- DOI: 10.1080/10409238.2017.1409191
Protein depalmitoylases
Abstract
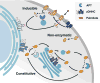
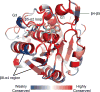
Protein depalmitoylation describes the removal of thioester-linked long chain fatty acids from cysteine residues in proteins. For many S-palmitoylated proteins, this process is promoted by acyl protein thioesterase enzymes, which catalyze thioester hydrolysis to solubilize and displace substrate proteins from membranes. The closely related enzymes acyl protein thioesterase 1 (APT1; LYPLA1) and acyl protein thioesterase 2 (APT2; LYPLA2) were initially identified from biochemical assays as G protein depalmitoylases, yet later were shown to accept a number of S-palmitoylated protein and phospholipid substrates. Leveraging the development of isoform-selective APT inhibitors, several studies report distinct roles for APT enzymes in growth factor and hormonal signaling. Recent crystal structures of APT1 and APT2 reveal convergent acyl binding channels, suggesting additional factors beyond acyl chain recognition mediate substrate selection. In addition to APT enzymes, the ABHD17 family of hydrolases contributes to the depalmitoylation of Ras-family GTPases and synaptic proteins. Overall, enzymatic depalmitoylation ensures efficient membrane targeting by balancing the palmitoylation cycle, and may play additional roles in signaling, growth, and cell organization. In this review, we provide a perspective on the biochemical, structural, and cellular analysis of protein depalmitoylases, and outline opportunities for future studies of systems-wide analysis of protein depalmitoylation.
Keywords: Palmitoylation; inhibitor; post-translational modification; serine hydrolase; thioesterase.
Conflict of interest statement
The authors disclose no conflict of interest.
Figures
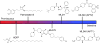

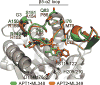


Similar articles
-
Dynamic palmitoylation links cytosol-membrane shuttling of acyl-protein thioesterase-1 and acyl-protein thioesterase-2 with that of proto-oncogene H-ras product and growth-associated protein-43.J Biol Chem. 2013 Mar 29;288(13):9112-25. doi: 10.1074/jbc.M112.421073. Epub 2013 Feb 8. J Biol Chem. 2013. PMID: 23396970 Free PMC article.
-
ABHD17 proteins are novel protein depalmitoylases that regulate N-Ras palmitate turnover and subcellular localization.Elife. 2015 Dec 23;4:e11306. doi: 10.7554/eLife.11306. Elife. 2015. PMID: 26701913 Free PMC article.
-
Enzymatic protein depalmitoylation by acyl protein thioesterases.Biochem Soc Trans. 2015 Apr;43(2):193-8. doi: 10.1042/BST20140235. Biochem Soc Trans. 2015. PMID: 25849916 Review.
-
Molecular Mechanism for Isoform-Selective Inhibition of Acyl Protein Thioesterases 1 and 2 (APT1 and APT2).ACS Chem Biol. 2016 Dec 16;11(12):3374-3382. doi: 10.1021/acschembio.6b00720. Epub 2016 Oct 31. ACS Chem Biol. 2016. PMID: 27748579 Free PMC article.
-
Depalmitoylation and cell physiology: APT1 as a mediator of metabolic signals.Am J Physiol Cell Physiol. 2024 Apr 1;326(4):C1034-C1041. doi: 10.1152/ajpcell.00542.2023. Epub 2024 Feb 12. Am J Physiol Cell Physiol. 2024. PMID: 38344800 Free PMC article. Review.
Cited by
-
Discovery and Characterization of IFITM S-Palmitoylation.Viruses. 2023 Nov 28;15(12):2329. doi: 10.3390/v15122329. Viruses. 2023. PMID: 38140570 Free PMC article. Review.
-
Synthetic Fluorogenic Peptides Reveal Dynamic Substrate Specificity of Depalmitoylases.Cell Chem Biol. 2019 Jan 17;26(1):35-47.e7. doi: 10.1016/j.chembiol.2018.10.005. Epub 2018 Nov 1. Cell Chem Biol. 2019. PMID: 30393067 Free PMC article.
-
Emerging roles of protein palmitoylation and its modifying enzymes in cancer cell signal transduction and cancer therapy.Int J Biol Sci. 2022 May 9;18(8):3447-3457. doi: 10.7150/ijbs.72244. eCollection 2022. Int J Biol Sci. 2022. PMID: 35637973 Free PMC article. Review.
-
GABAAR-PPT1 palmitoylation homeostasis controls synaptic transmission and circuitry oscillation.Transl Psychiatry. 2024 Dec 18;14(1):488. doi: 10.1038/s41398-024-03206-1. Transl Psychiatry. 2024. PMID: 39695089 Free PMC article.
-
An S-acylated N-terminus and a conserved loop regulate the activity of the ABHD17 deacylase.J Cell Biol. 2025 Apr 7;224(4):e202405042. doi: 10.1083/jcb.202405042. Epub 2025 Feb 14. J Cell Biol. 2025. PMID: 39951021
References
-
- Adibekian A, Martin BR, Chang JW, Hsu KL, Tsuboi K, Bachovchin DA, Speers AE, Brown SJ, Spicer T, Fernandez-Vega V, et al. Probe Reports from the NIH Molecular Libraries Program. Bethesda (MD): 2010a. Characterization of a Selective, Reversible Inhibitor of Lysophospholipase 1 (LYPLA1) - PubMed
-
- Adibekian A, Martin BR, Chang JW, Hsu KL, Tsuboi K, Bachovchin DA, Speers AE, Brown SJ, Spicer T, Fernandez-Vega V, et al. Probe Reports from the NIH Molecular Libraries Program. Bethesda (MD): 2010b. Characterization of a Selective, Reversible Inhibitor of Lysophospholipase 2 (LYPLA2) - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
