Live-cell mapping of organelle-associated RNAs via proximity biotinylation combined with protein-RNA crosslinking
- PMID: 29239719
- PMCID: PMC5730372
- DOI: 10.7554/eLife.29224
Live-cell mapping of organelle-associated RNAs via proximity biotinylation combined with protein-RNA crosslinking
Abstract
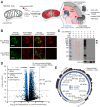
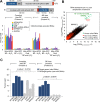
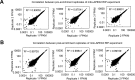
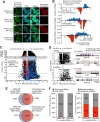
The spatial organization of RNA within cells is a crucial factor influencing a wide range of biological functions throughout all kingdoms of life. However, a general understanding of RNA localization has been hindered by a lack of simple, high-throughput methods for mapping the transcriptomes of subcellular compartments. Here, we develop such a method, termed APEX-RIP, which combines peroxidase-catalyzed, spatially restricted in situ protein biotinylation with RNA-protein chemical crosslinking. We demonstrate that, using a single protocol, APEX-RIP can isolate RNAs from a variety of subcellular compartments, including the mitochondrial matrix, nucleus, cytosol, and endoplasmic reticulum (ER), with specificity and sensitivity that rival or exceed those of conventional approaches. We further identify candidate RNAs localized to mitochondria-ER junctions and nuclear lamina, two compartments that are recalcitrant to classical biochemical purification. Since APEX-RIP is simple, versatile, and does not require special instrumentation, we envision its broad application in a variety of biological contexts.
Keywords: APEX2; Horseradish peroxidase; RNA localization; RNA sequencing; biochemistry; cell biology; human; peroxidase; subcellular transcriptome.
Conflict of interest statement
No competing interests declared.
Figures


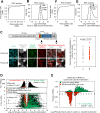



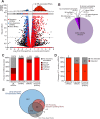
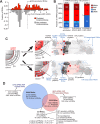
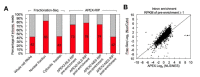
References
-
- Adiconis X, Borges-Rivera D, Satija R, DeLuca DS, Busby MA, Berlin AM, Sivachenko A, Thompson DA, Wysoker A, Fennell T, Gnirke A, Pochet N, Regev A, Levin JZ. Comparative analysis of RNA sequencing methods for degraded or low-input samples. Nature Methods. 2013;10:623–629. doi: 10.1038/nmeth.2483. - DOI - PMC - PubMed
-
- Alán L, Zelenka J, Ježek J, Dlasková A, Ježek P. Fluorescent in situ hybridization of mitochondrial DNA and RNA. Acta Biochimica Polonica. 2010;57:403–408. - PubMed
-
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene Ontology: tool for the unification of biology. Nature Genetics. 2000;25:25–29. doi: 10.1038/75556. - DOI - PMC - PubMed
-
- Batish M, Raj A, Tyagi S. RNA Detection and Visualization. 2011. Single Molecule Imaging of RNA; pp. 3–13. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

