Single-cell RNA sequencing reveals intrinsic and extrinsic regulatory heterogeneity in yeast responding to stress
- PMID: 29240790
- PMCID: PMC5746276
- DOI: 10.1371/journal.pbio.2004050
Single-cell RNA sequencing reveals intrinsic and extrinsic regulatory heterogeneity in yeast responding to stress
Abstract
From bacteria to humans, individual cells within isogenic populations can show significant variation in stress tolerance, but the nature of this heterogeneity is not clear. To investigate this, we used single-cell RNA sequencing to quantify transcript heterogeneity in single Saccharomyces cerevisiae cells treated with and without salt stress to explore population variation and identify cellular covariates that influence the stress-responsive transcriptome. Leveraging the extensive knowledge of yeast transcriptional regulation, we uncovered significant regulatory variation in individual yeast cells, both before and after stress. We also discovered that a subset of cells appears to decouple expression of ribosomal protein genes from the environmental stress response in a manner partly correlated with the cell cycle but unrelated to the yeast ultradian metabolic cycle. Live-cell imaging of cells expressing pairs of fluorescent regulators, including the transcription factor Msn2 with Dot6, Sfp1, or MAP kinase Hog1, revealed both coordinated and decoupled nucleocytoplasmic shuttling. Together with transcriptomic analysis, our results suggest that cells maintain a cellular filter against decoupled bursts of transcription factor activation but mount a stress response upon coordinated regulation, even in a subset of unstressed cells.
Conflict of interest statement
I have read the journal's policy and the authors of this manuscript have the following competing interests: SRQ is a co-founder of Fluidigm, whose technology was used as part of this project.
Figures

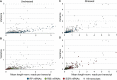



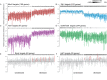

References
-
- Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science (New York, NY. 2004;305(5690):1622–5. - PubMed
-
- Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141(1):69–80. doi: 10.1016/j.cell.2010.02.027 - DOI - PMC - PubMed
-
- Roesch A, Fukunaga-Kalabis M, Schmidt EC, Zabierowski SE, Brafford PA, Vultur A, et al. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell. 2010;141(4):583–94. doi: 10.1016/j.cell.2010.04.020 - DOI - PMC - PubMed
-
- Shaffer SM, Dunagin MC, Torborg SR, Torre EA, Emert B, Krepler C, et al. Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature. 2017;546(7658):431–5. doi: 10.1038/nature22794 - DOI - PMC - PubMed
-
- Andrusiak K. Adapting S. cerevisiae Chemical Genomics for Identifying the Modes of Action of Natural Compounds Toronto: University of Toronto; 2012.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

