Machine learning for classifying tuberculosis drug-resistance from DNA sequencing data
- PMID: 29240876
- PMCID: PMC5946815
- DOI: 10.1093/bioinformatics/btx801
Machine learning for classifying tuberculosis drug-resistance from DNA sequencing data
Abstract
Motivation: Correct and rapid determination of Mycobacterium tuberculosis (MTB) resistance against available tuberculosis (TB) drugs is essential for the control and management of TB. Conventional molecular diagnostic test assumes that the presence of any well-studied single nucleotide polymorphisms is sufficient to cause resistance, which yields low sensitivity for resistance classification.
Summary: Given the availability of DNA sequencing data from MTB, we developed machine learning models for a cohort of 1839 UK bacterial isolates to classify MTB resistance against eight anti-TB drugs (isoniazid, rifampicin, ethambutol, pyrazinamide, ciprofloxacin, moxifloxacin, ofloxacin, streptomycin) and to classify multi-drug resistance.
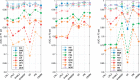
Results: Compared to previous rules-based approach, the sensitivities from the best-performing models increased by 2-4% for isoniazid, rifampicin and ethambutol to 97% (P < 0.01), respectively; for ciprofloxacin and multi-drug resistant TB, they increased to 96%. For moxifloxacin and ofloxacin, sensitivities increased by 12 and 15% from 83 and 81% based on existing known resistance alleles to 95% and 96% (P < 0.01), respectively. Particularly, our models improved sensitivities compared to the previous rules-based approach by 15 and 24% to 84 and 87% for pyrazinamide and streptomycin (P < 0.01), respectively. The best-performing models increase the area-under-the-ROC curve by 10% for pyrazinamide and streptomycin (P < 0.01), and 4-8% for other drugs (P < 0.01).
Availability and implementation: The details of source code are provided at http://www.robots.ox.ac.uk/~davidc/code.php.
Contact: david.clifton@eng.ox.ac.uk.
Supplementary information: Supplementary data are available at Bioinformatics online.
Figures


References
-
- Ando H. et al. (2014) A silent mutation in maba confers isoniazid resistance on Mycobacterium tuberculosis. Mol. Microbiol., 91, 538–547. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

