Associations between infant fungal and bacterial dysbiosis and childhood atopic wheeze in a nonindustrialized setting
- PMID: 29241587
- PMCID: PMC6075469
- DOI: 10.1016/j.jaci.2017.08.041
Associations between infant fungal and bacterial dysbiosis and childhood atopic wheeze in a nonindustrialized setting
Abstract
Background: Asthma is the most prevalent chronic disease of childhood. Recently, we identified a critical window early in the life of both mice and Canadian infants during which gut microbial changes (dysbiosis) affect asthma development. Given geographic differences in human gut microbiota worldwide, we studied the effects of gut microbial dysbiosis on atopic wheeze in a population living in a distinct developing world environment.
Objective: We sought to determine whether microbial alterations in early infancy are associated with the development of atopic wheeze in a nonindustrialized setting.
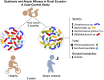
Methods: We conducted a case-control study nested within a birth cohort from rural Ecuador in which we identified 27 children with atopic wheeze and 70 healthy control subjects at 5 years of age. We analyzed bacterial and eukaryotic gut microbiota in stool samples collected at 3 months of age using 16S and 18S sequencing. Bacterial metagenomes were predicted from 16S rRNA data by using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States and categorized by function with Kyoto Encyclopedia of Genes and Genomes ontology. Concentrations of fecal short-chain fatty acids were determined by using gas chromatography.
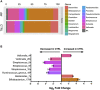
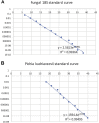
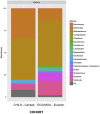
Results: As previously observed in Canadian infants, microbial dysbiosis at 3 months of age was associated with later development of atopic wheeze. However, the dysbiosis in Ecuadorian babies involved different bacterial taxa, was more pronounced, and also involved several fungal taxa. Predicted metagenomic analysis emphasized significant dysbiosis-associated differences in genes involved in carbohydrate and taurine metabolism. Levels of the fecal short-chain fatty acids acetate and caproate were reduced and increased, respectively, in the 3-month stool samples of children who went on to have atopic wheeze.
Conclusions: Our findings support the importance of fungal and bacterial microbiota during the first 100 days of life on the development of atopic wheeze and provide additional support for considering modulation of the gut microbiome as a primary asthma prevention strategy.
Keywords: Asthma; atopy; gut microbiome; mycobiome; nonindustrialized setting; short-chain fatty acids; wheeze.
Copyright © 2017 The Authors. Published by Elsevier Inc. All rights reserved.
Figures






References
-
- Asher I., Pearce N. Global burden of asthma among children. Int J Tuberc Lung Dis. 2014;18:1269–1278. - PubMed
-
- Russell S.L., Gold M.J., Reynolds L.A., Willing B.P., Dimitriu P., Thorson L. Perinatal antibiotic-induced shifts in gut microbiota have differential effects on inflammatory lung diseases. J Allergy Clin Immunol. 2015;135:100–109. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

