Rational library design by functional CDR resampling
- PMID: 29242049
- PMCID: PMC5995609
- DOI: 10.1016/j.nbt.2017.12.005
Rational library design by functional CDR resampling
Abstract
Successful antibody discovery relies on diversified libraries, where two aspects are implied, namely the absolute number of unique clones and the percentage of functional clones. Instead of pursuing the absolute quantity thresholded by current display technology, we have sought to maximize the effective diversity by improving functional clone percentage. With the combined effort of bioinformatics, structural biology, molecular immunology and phage display technology, we devised a bioinformatic pipeline to construct and validate libraries via combinatorial assembly of sequences from a database of experimentally validated antibodies. Furthermore, we showed that the libraries constructed as such yielded a significantly increased success rate against different antigen types and generated over 20-fold more unique hits per targets compared with libraries based on traditional degenerate nucleotide methods. Our study indicated that predefined CDR sequences with optimized CDR-framework compatibility could be a productive direction of functional library construction for in vitro antibody development.
Copyright © 2017 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declared no conflict of interests.
Figures




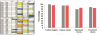


References
-
- da Silva MC, Del Sarto RP, Lucena WA, Rigden DJ, Teixeira FR, Bezerra Cde A, et al. Employing in vitro directed molecular evolution for the selection of alpha-amylase variant inhibitors with activity toward cotton boll weevil enzyme. J Biotechnol. 2013;167:377–385. - PubMed
-
- Riano-Umbarila L, Juarez-Gonzalez VR, Olamendi-Portugal T, Ortiz-Leon M, Possani LD, Becerril B. A strategy for the generation of specific human antibodies by directed evolution and phage display, An example of a single-chain antibody fragment that neutralizes a major component of scorpion venom. FEBS J. 2005;272:2591–2601. - PubMed
-
- Xu L, Aha P, Gu K, Kuimelis RG, Kurz M, Lam T, et al. Directed evolution of high-affinity antibody mimics using mRNA display. Chem Biol. 2002;9:933–942. - PubMed
-
- Juarez-Gonzalez VR, Riano-Umbarila L, Quintero-Hernandez V, Olamendi-Portugal T, Ortiz-Leon M, Ortiz E, et al. Directed evolution, phage display and combination of evolved mutants: a strategy to recover the neutralization properties of the scFv version of BCF2 a neutralizing monoclonal antibody specific to scorpion toxin Cn2. J Mol Biol. 2005;346:1287–1297. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

