The ER membrane protein complex is a transmembrane domain insertase
- PMID: 29242231
- PMCID: PMC5788257
- DOI: 10.1126/science.aao3099
The ER membrane protein complex is a transmembrane domain insertase
Abstract
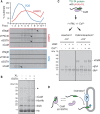
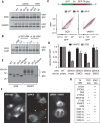
Insertion of proteins into membranes is an essential cellular process. The extensive biophysical and topological diversity of membrane proteins necessitates multiple insertion pathways that remain incompletely defined. Here we found that known membrane insertion pathways fail to effectively engage tail-anchored membrane proteins with moderately hydrophobic transmembrane domains. These proteins are instead shielded in the cytosol by calmodulin. Dynamic release from calmodulin allowed sampling of the endoplasmic reticulum (ER), where the conserved ER membrane protein complex (EMC) was shown to be essential for efficient insertion in vitro and in cells. Purified EMC in synthetic liposomes catalyzed the insertion of its substrates in a reconstituted system. Thus, EMC is a transmembrane domain insertase, a function that may explain its widely pleiotropic membrane-associated phenotypes across organisms.
Copyright © 2018, American Association for the Advancement of Science.
Figures


Comment in
-
Oxa1 Superfamily: New Members Found in the ER.Trends Biochem Sci. 2018 Mar;43(3):151-153. doi: 10.1016/j.tibs.2017.12.005. Epub 2018 Jan 12. Trends Biochem Sci. 2018. PMID: 29310909
-
Complexity in targeting membrane proteins.Science. 2018 Jan 26;359(6374):390-391. doi: 10.1126/science.aar5992. Science. 2018. PMID: 29371455 No abstract available.
References
-
- Kalbfleisch T, Cambon A, Wattenberg B. W, Traffic 8, 1687–1694 (2007). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

