Gut microbiome: a new player in gastrointestinal disease
- PMID: 29243124
- PMCID: PMC5849673
- DOI: 10.1007/s00428-017-2277-x
Gut microbiome: a new player in gastrointestinal disease
Abstract
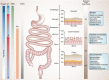
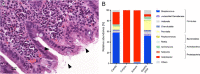
The gastrointestinal (GI) tract harbors a diverse and host-specific gut microbial community. Whereas host-microbe interactions are based on homeostasis and mutualism, the microbiome also contributes to disease development. In this review, we summarize recent findings connecting the GI microbiome with GI disease. Starting with a description of biochemical factors shaping microbial compositions in each gut segment along the longitudinal axis, improved histological techniques enabling high resolution visualization of the spatial microbiome structure are highlighted. Subsequently, inflammatory and neoplastic diseases of the esophagus, stomach, and small and large intestines are discussed and the respective changes in microbiome compositions summarized. Finally, approaches aiming to restore disturbed microbiome compositions thereby promoting health are discussed.
Keywords: Carcinogenesis; Dysbiosis; Fecal microbiome transplantation; Gut microbiome; Inflammation; Spatial microbiome organization.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures


References
-
- Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med. 2016;375(24):2369–2379. - PubMed
-
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Doré J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, MetaHIT Consortium. Bork P, Ehrlich SD, Wang J. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

