Neurodegenerative Disease Proteinopathies Are Connected to Distinct Histone Post-translational Modification Landscapes
- PMID: 29243911
- PMCID: PMC5906139
- DOI: 10.1021/acschemneuro.7b00297
Neurodegenerative Disease Proteinopathies Are Connected to Distinct Histone Post-translational Modification Landscapes
Abstract
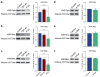
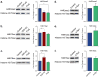
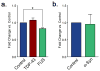
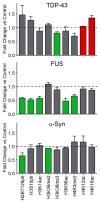
Amyotrophic lateral sclerosis (ALS) and Parkinson's disease (PD) are devastating neurodegenerative diseases involving the progressive degeneration of neurons. No cure is available for patients diagnosed with these diseases. A prominent feature of both ALS and PD is the accumulation of protein inclusions in the cytoplasm of degenerating neurons; however, the particular proteins constituting these inclusions vary: the RNA-binding proteins TDP-43 and FUS are most notable in ALS, while α-synuclein aggregates into Lewy bodies in PD. In both diseases, genetic causes fail to explain the occurrence of a large proportion of cases, and thus, both are considered mostly sporadic. Despite mounting evidence for a possible role of epigenetics in the occurrence and progression of ALS and PD, epigenetic mechanisms in the context of these diseases remain mostly unexplored. Here we comprehensively delineate histone post-translational modification (PTM) profiles in ALS and PD yeast proteinopathy models. Remarkably, we find distinct changes in histone modification profiles for each. We detect the most striking changes in the context of FUS aggregation: changes in several histone marks support a global decrease in gene transcription. We also detect more modest changes in histone modifications in cells overexpressing TDP-43 or α-synuclein. Our results highlight a great need for the inclusion of epigenetic mechanisms in the study of neurodegeneration. We hope our work will pave the way for the discovery of more effective therapies to treat patients suffering from ALS, PD, and other neurodegenerative diseases.
Keywords: ALS; FUS; Parkinson’s disease; TDP-43; epigenetics; histone modifications; α-synuclein.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


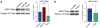
References
-
- Beitz JM. Parkinson’s disease: a review. Front Biosci, Scholar Ed. 2014;S6:65–74. - PubMed
-
- Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A, Kanagaraj AP, Carter R, Boylan KB, Wojtas AM, Rademakers R, Pinkus JL, Greenberg SA, Trojanowski JQ, Traynor BJ, Smith BN, Topp S, Gkazi AS, Miller J, Shaw CE, Kottlors M, Kirschner J, Pestronk A, Li YR, Ford AF, Gitler AD, Benatar M, King OD, Kimonis VE, Ross ED, Weihl CC, Shorter J, Taylor JP. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495:467–473. - PMC - PubMed
-
- Poewe W, Seppi K, Tanner CM, Halliday GM, Brundin P, Volkmann J, Schrag AE, Lang AE. Parkinson disease. Nature reviews Disease primers. 2017;3:17013. - PubMed
-
- Robberecht W, Philips T. The changing scene of amyotrophic lateral sclerosis. Nat Rev Neurosci. 2013;14:248–264. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

