RUNX1 promote invasiveness in pancreatic ductal adenocarcinoma through regulating miR-93
- PMID: 29245924
- PMCID: PMC5725115
- DOI: 10.18632/oncotarget.20433
RUNX1 promote invasiveness in pancreatic ductal adenocarcinoma through regulating miR-93
Abstract
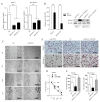
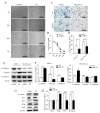
Runt-related transcription factor 1(RUNX1), a key factor in hematopoiesis that mediates specification and homeostasis of hematopoietic stem and progenitor cells (HSPCs), is also overexpressed in several solid human cancers, and correlated with tumor progression. However, the expression and function of RUNX1 in pancreatic ductal adenocarcinoma were still unclear. Here, we show that RUNX1 is highly expressed in pancreatic adenocarcinoma tissues and knocking down of RUNX1 attenuated aggressiveness in pancreatic cell lines. Moreover, we found that RUNX1 could negatively regulate the expression of miR-93. Bioinformatics method showed that there are two binding sites in the the promotor region of miR-93 precursor and through ChIP-qPCR and firefly luciferase reporter assay, we vertified that these two binding sites each have transcriptive activity in one pancreatic cell lines. This result supported our presumption that RUNX1 regulate miR-93 through binding to the promotor region of miR-93. Besides, the expression and function of miR-93 is quite the opposite, miR-93 overexpression suppresses migration and invasiveness in pancreatic cell lines supporting that RUNX1 negatively regulated miR-93. Our findings provided evidence regarding the role of RUNX1 as an oncogene through the inhibition of miR-93. Targeting RUNX1 can be a potential therapeutic strategy in pancreatic cancer.
Keywords: HMGA2 (High mobility group AT-hook 2); RUNX1; miR-93; pancreatic ductal adenocarcinoma (PDAC); tumor progression.
Conflict of interest statement
CONFLICTS OF INTEREST The authors declare no competing financial interests.
Figures




References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. https://doi.org/10.3322/caac.21332. - DOI - PubMed
-
- Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, Rosso S, Coebergh JW, Comber H, Forman D, Bray F. Cancer incidence and mortality patterns in Europe: estimates for 40 countries in 2012. Eur J Cancer. 2013;49:1374–403. https://doi.org/10.1016/j.ejca.2012.12.027. - DOI - PubMed
-
- Chen X, Yang H, Zhou X, Zhang L, Lu X. MiR-93 Targeting EphA4 Promotes Neurite Outgrowth from Spinal Cord Neurons. J Mol Neurosci. 2016;58:517–24. https://doi.org/10.1007/s12031-015-0709-0. - DOI - PubMed
-
- Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med. 2014;371:1039–49. https://doi.org/10.1056/NEJMra1404198. - DOI - PubMed
-
- Kamisawa T, Wood LD, Itoi T, Takaori K. Pancreatic cancer. The Lancet. 388:73–85. https://doi.org/10.1016/S0140-6736(16)00141-0. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

