Canary: an atomic pipeline for clinical amplicon assays
- PMID: 29246107
- PMCID: PMC5732437
- DOI: 10.1186/s12859-017-1950-z
Canary: an atomic pipeline for clinical amplicon assays
Abstract
Background: High throughput sequencing requires bioinformatics pipelines to process large volumes of data into meaningful variants that can be translated into a clinical report. These pipelines often suffer from a number of shortcomings: they lack robustness and have many components written in multiple languages, each with a variety of resource requirements. Pipeline components must be linked together with a workflow system to achieve the processing of FASTQ files through to a VCF file of variants. Crafting these pipelines requires considerable bioinformatics and IT skills beyond the reach of many clinical laboratories.
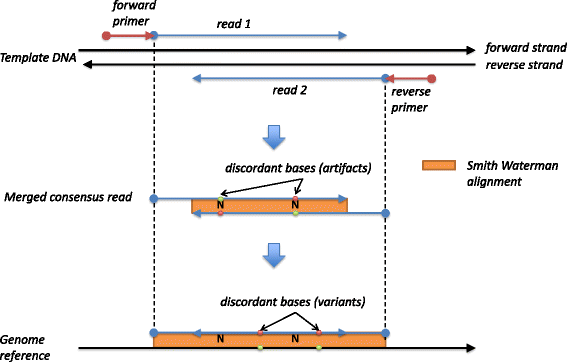
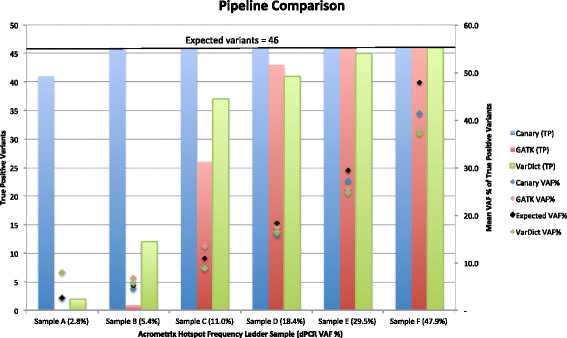
Results: Here we present Canary, a single program that can be run on a laptop, which takes FASTQ files from amplicon assays through to an annotated VCF file ready for clinical analysis. Canary can be installed and run with a single command using Docker containerization or run as a single JAR file on a wide range of platforms. Although it is a single utility, Canary performs all the functions present in more complex and unwieldy pipelines. All variants identified by Canary are 3' shifted and represented in their most parsimonious form to provide a consistent nomenclature, irrespective of sequencing variation. Further, proximate in-phase variants are represented as a single HGVS 'delins' variant. This allows for correct nomenclature and consequences to be ascribed to complex multi-nucleotide polymorphisms (MNPs), which are otherwise difficult to represent and interpret. Variants can also be annotated with hundreds of attributes sourced from MyVariant.info to give up to date details on pathogenicity, population statistics and in-silico predictors.
Conclusions: Canary has been used at the Peter MacCallum Cancer Centre in Melbourne for the last 2 years for the processing of clinical sequencing data. By encapsulating clinical features in a single, easily installed executable, Canary makes sequencing more accessible to all pathology laboratories. Canary is available for download as source or a Docker image at https://github.com/PapenfussLab/Canary under a GPL-3.0 License.
Keywords: Amplicon; Canary; Clinical diagnostics; PathOS; Pipelines; Targeted sequencing; Variant calling.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

References
-
- Docker. Docker containerisation site, http:/http://www.docker.com. Accessed 29 Nov 2017.
-
- Illumina. https://basespace.illumina.com.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

