Distinct pH dependencies of Na+/K+ selectivity at the two faces of Na,K-ATPase
- PMID: 29247005
- PMCID: PMC5808778
- DOI: 10.1074/jbc.RA117.000700
Distinct pH dependencies of Na+/K+ selectivity at the two faces of Na,K-ATPase
Abstract
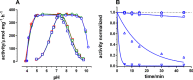
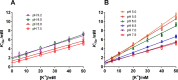
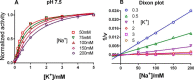
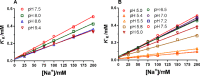
The sodium pump (Na,K-ATPase) in animal cells is vital for actively maintaining ATP hydrolysis-powered Na+ and K+ electrochemical gradients across the cell membrane. These ion gradients drive co- and countertransport and are critical for establishing the membrane potential. It has been an enigma how Na,K-ATPase discriminates between Na+ and K+, despite the pumped ion on each side being at a lower concentration than the other ion. Recent crystal structures of analogs of the intermediate conformations E2·Pi·2K+ and Na+-bound E1∼P·ADP suggest that the dimensions of the respective binding sites in Na,K-ATPase are crucial in determining its selectivity. Here, we found that the selectivity at each membrane face is pH-dependent and that this dependence is unique for each face. Most notable was a strong increase in the specific affinity for K+ at the extracellular face (i.e. E2 conformation) as the pH is lowered from 7.5 to 5. We also observed a smaller increase in affinity for K+ on the cytoplasmic side (E1 conformation), which reduced the selectivity for Na+ Theoretical analysis of the pKa values of ion-coordinating acidic amino acid residues suggested that the face-specific pH dependences and Na+/K+ selectivities may arise from the protonation or ionization of key residues. The increase in K+ selectivity at low pH on the cytoplasmic face, for instance, appeared to be associated with Asp808 protonation. We conclude that changes in the ionization state of coordinating residues in Na,K-ATPase could contribute to altering face-specific ion selectivity.
Keywords: ATPase; E1/E2 conformations; enzyme kinetics; enzyme mechanism; membrane protein; membrane transport; pH; pKa; protonation of binding sites.
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interests with the contents of this article
Figures





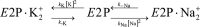




References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

