The transcriptome of Mycobacterium tuberculosis in a lipid-rich dormancy model through RNAseq analysis
- PMID: 29247215
- PMCID: PMC5732278
- DOI: 10.1038/s41598-017-17751-x
The transcriptome of Mycobacterium tuberculosis in a lipid-rich dormancy model through RNAseq analysis
Abstract
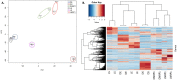
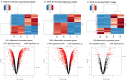
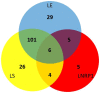
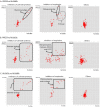
Tuberculosis (TB) is currently the number one killer among infectious diseases worldwide. Lipids are abundant molecules during the infectious cycle of Mycobacterium tuberculosis (Mtb) and studies better mimicking its actual metabolic state during pathogenesis are needed. Though most studies have focused on the mycobacterial lipid metabolism under standard culture conditions, little is known about the transcriptome of Mtb in a lipid environment. Here we determined the transcriptome of Mtb H37Rv in a lipid-rich environment (cholesterol and fatty acid) under aerobic and hypoxic conditions, using RNAseq. Lipids significantly induced the expression of 368 genes. A main core lipid response was observed involving efflux systems, iron caption and sulfur reduction. In co-expression with ncRNAs and other genes discussed below, may act coordinately to prepare the machinery conferring drug tolerance and increasing a persistent population. Our findings could be useful to tag relevant pathways for the development of new drugs, vaccines and new strategies to control TB.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
-
- WHO. Global tuberculosis report 2016. Geneva: World Health Organization Available at: http://www.who.int/tb/about/ (2016).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

