Nudt21 Controls Cell Fate by Connecting Alternative Polyadenylation to Chromatin Signaling
- PMID: 29249356
- PMCID: PMC5766360
- DOI: 10.1016/j.cell.2017.11.023
Nudt21 Controls Cell Fate by Connecting Alternative Polyadenylation to Chromatin Signaling
Erratum in
-
Nudt21 Controls Cell Fate by Connecting Alternative Polyadenylation to Chromatin Signaling.Cell. 2018 Jan 25;172(3):629-631. doi: 10.1016/j.cell.2017.12.035. Cell. 2018. PMID: 29373832 Free PMC article. No abstract available.
Abstract
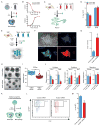
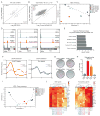
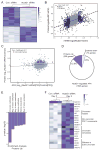
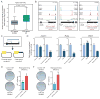
Cell fate transitions involve rapid gene expression changes and global chromatin remodeling, yet the underlying regulatory pathways remain incompletely understood. Here, we identified the RNA-processing factor Nudt21 as a novel regulator of cell fate change using transcription-factor-induced reprogramming as a screening assay. Suppression of Nudt21 enhanced the generation of induced pluripotent stem cells, facilitated transdifferentiation into trophoblast stem cells, and impaired differentiation of myeloid precursors and embryonic stem cells, suggesting a broader role for Nudt21 in cell fate change. We show that Nudt21 directs differential polyadenylation of over 1,500 transcripts in cells acquiring pluripotency, although only a fraction changed protein levels. Remarkably, these proteins were strongly enriched for chromatin regulators, and their suppression neutralized the effect of Nudt21 during reprogramming. Collectively, our data uncover Nudt21 as a novel post-transcriptional regulator of cell fate and establish a direct, previously unappreciated link between alternative polyadenylation and chromatin signaling.
Keywords: Alternative polyadenylation; chromatin; embryonic stem cells; epigenetic regulation; induced pluripotent stem cells; induced trophoblast stem cells; microRNA; pluripotency; reprogramming; transdifferentiation.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures



References
-
- Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nature biotechnology. 2006;24:1285–1292. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

