JAK/STAT3-Regulated Fatty Acid β-Oxidation Is Critical for Breast Cancer Stem Cell Self-Renewal and Chemoresistance
- PMID: 29249690
- PMCID: PMC5777338
- DOI: 10.1016/j.cmet.2017.11.001
JAK/STAT3-Regulated Fatty Acid β-Oxidation Is Critical for Breast Cancer Stem Cell Self-Renewal and Chemoresistance
Erratum in
-
JAK/STAT3-Regulated Fatty Acid β-Oxidation Is Critical for Breast Cancer Stem Cell Self-Renewal and Chemoresistance.Cell Metab. 2018 Jun 5;27(6):1357. doi: 10.1016/j.cmet.2018.04.018. Cell Metab. 2018. PMID: 29874570 Free PMC article. No abstract available.
Abstract
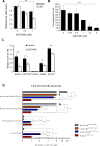
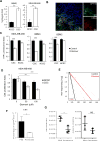
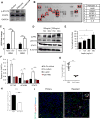
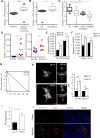
Cancer stem cells (CSCs) are critical for cancer progression and chemoresistance. How lipid metabolism regulates CSCs and chemoresistance remains elusive. Here, we demonstrate that JAK/STAT3 regulates lipid metabolism, which promotes breast CSCs (BCSCs) and cancer chemoresistance. Inhibiting JAK/STAT3 blocks BCSC self-renewal and expression of diverse lipid metabolic genes, including carnitine palmitoyltransferase 1B (CPT1B), which encodes the critical enzyme for fatty acid β-oxidation (FAO). Moreover, mammary-adipocyte-derived leptin upregulates STAT3-induced CPT1B expression and FAO activity in BCSCs. Human breast-cancer-derived data suggest that the STAT3-CPT1B-FAO pathway promotes cancer cell stemness and chemoresistance. Blocking FAO and/or leptin re-sensitizes them to chemotherapy and inhibits BCSCs in mouse breast tumors in vivo. We identify a critical pathway for BCSC maintenance and breast cancer chemoresistance.
Keywords: JAK/STAT3; breast cancer stem cell; chemoresistance; fatty acid β-oxidation; leptin.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures


Comment in
-
Blocking leptin-STAT3 axis-induced fatty acid oxidation: A novel approach to activate CD8+ T effector cells in breast cancer.Thorac Cancer. 2020 Dec;11(12):3422-3424. doi: 10.1111/1759-7714.13691. Epub 2020 Oct 16. Thorac Cancer. 2020. PMID: 33063947 Free PMC article. No abstract available.
References
-
- Abumrad NA, Elmaghrabi MR, Amri EZ, Lopez E, Grimaldi PA. Cloning of a Rat Adipocyte Membrane-Protein Implicated in Binding or Transport of Long-Chain Fatty-Acids That Is Induced during Preadipocyte Differentiation - Homology with Human Cd36. Journal of Biological Chemistry. 1993;268:17665–17668. - PubMed
-
- Bozza PT, Viola JP. Lipid droplets in inflammation and cancer. Prostaglandins, leukotrienes, and essential fatty acids. 2010;82:243–250. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

