The Microbiome of Potentially Malignant Oral Leukoplakia Exhibits Enrichment for Fusobacterium, Leptotrichia, Campylobacter, and Rothia Species
- PMID: 29250055
- PMCID: PMC5717034
- DOI: 10.3389/fmicb.2017.02391
The Microbiome of Potentially Malignant Oral Leukoplakia Exhibits Enrichment for Fusobacterium, Leptotrichia, Campylobacter, and Rothia Species
Abstract
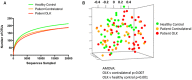
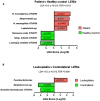
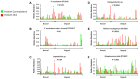
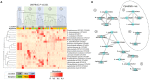
Oral leukoplakia presents as a white patch on the oral mucosa and is recognized as having significant malignant potential. Although colonization of these patches with Candida albicans is common, little is known about the bacterial microbiota of these patches. In the current study we analyzed the microbiome of oral leukoplakia in 36 patients compared to healthy mucosal tissue from the same patients and healthy control subjects to determine if specific microbial enrichments could be identified early in the malignant process that could play a role in the progression of the disease. This was carried out by sequence analysis of the V1-V2 region of the bacterial 16S rRNA gene using the Illumina MiSeq. Oral leukoplakia exhibited increased abundance of Fusobacteria and reduced levels of Firmicutes (Metastats P < 0.01). Candida colonization was also more prevalent in leukoplakia patients relative to healthy controls (P = 0.025). Bacterial colonization patterns on oral leukoplakia were highly variable and five distinct bacterial clusters were discerned. These clusters exhibited co-occurrence of Fusobacterium, Leptotrichia, and Campylobacter species (Pearson P < 0.01), which is strikingly similar to the microbial co-occurrence patterns observed on colorectal cancers (Warren et al., 2013). Increased abundance of the acetaldehydogenic microorganism Rothia mucilaginosa was also apparent on oral leukoplakias from lingual sites (P 0.0012). Severe dysplasia was associated with elevated levels of Leptotrichia spp. and Campylobacter concisus (P < 0.05). Oral leukoplakia exhibits an altered microbiota that has similarities to the microbiome of colorectal cancer.
Keywords: Campylobacter; Fusobacteria; Rothia mucilaginosa; microbiome; oral cancer; oral leukoplakia.
Figures

References
-
- Abdulrahim M. H., McManus B. A., Flint S. R., Coleman D. C. (2013). Genotyping Candida albicans from Candida leukoplakia and non-Candida leukoplakia shows no enrichment of multilocus sequence typing clades but enrichment of ABC genotype C in Candida leukoplakia. PLOS ONE 8:e73738. 10.1371/journal.pone.0073738 - DOI - PMC - PubMed
-
- Al-hebshi N. N., Nasher A. T., Maryoud M. Y., Homeida H. E., Chen T., Idris A. M., et al. (2017). Inflammatory bacteriome featuring Fusobacterium nucleatum and Pseudomonas aeruginosa identified in association with oral squamous cell carcinoma. Sci. Rep. 7:1834. 10.1038/s41598-017-02079-3 - DOI - PMC - PubMed
-
- Alnuaimi A. D., Wiesenfeld D., O’Brien-Simpson N. M., Reynolds E. C., McCullough M. J. (2015). Oral Candida colonization in oral cancer patients and its relationship with traditional risk factors of oral cancer: a matched case-control study. Oral Oncol. 51 139–145. 10.1016/j.oraloncology.2014.11.008 - DOI - PubMed
-
- Binder Gallimidi A., Fischman S., Revach B., Bulvik R., Maliutina A., Rubinstein A. M., et al. (2015). Periodontal pathogens Porphyromonas gingivalis and Fusobacterium nucleatum promote tumor progression in an oral-specific chemical carcinogenesis model. Oncotarget 6 22613–22623. 10.18632/oncotarget.4209 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

