Biochemical and Transcriptional Regulation of Membrane Lipid Metabolism in Maize Leaves under Low Temperature
- PMID: 29250095
- PMCID: PMC5714865
- DOI: 10.3389/fpls.2017.02053
Biochemical and Transcriptional Regulation of Membrane Lipid Metabolism in Maize Leaves under Low Temperature
Abstract
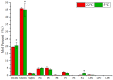
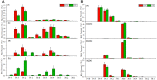
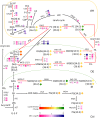
Membrane lipid modulation is one of the major strategies plants have developed for cold acclimation. In this study, a combined lipidomic and transcriptomic analysis was conducted, and the changes in glycerolipids contents and species, and transcriptional regulation of lipid metabolism in maize leaves under low temperature treatment (5°C) were investigated. The lipidomic analysis showed an increase in the phospholipid phosphatidic acid (PA) and a decrease in phosphatidylcholine (PC). And an increase in digalactosyldiacylglycerol and a decrease in monogalactosyldiacylglycerol of the galactolipid class. The results implied an enhanced turnover of PC to PA to serve as precursors for galactolipid synthesis under following low temperature treatment. The analysis of changes in abundance of various lipid molecular species suggested major alterations of different pathways of plastidic lipids synthesis in maize under cold treatment. The synchronous transcriptomic analysis revealed that genes involved in phospholipid and galactolipid synthesis pathways were significantly up-regulated, and a comprehensive gene-metabolite network was generated illustrating activated membrane lipids adjustment in maize leaves following cold treatment. This study will help to understand the regulation of glycerolipids metabolism at both biochemical and molecular biological levels in 18:3 plants and to decipher the roles played by lipid remodeling in cold response in major field crop maize.
Keywords: RNA-Seq; lipid metabolism; lipidome profiling; low temperature; maize.
Figures


References
-
- Beisson F., Koo A. J., Ruuska S., Schwender J., Pollard M., Thelen J. J., et al. (2003). Arabidopsis genes involved in acyl lipid metabolism. A 2003 census of the candidates, a study of the distribution of expressed sequence tags in organs, and a web-based database. Plant Physiol. 132 681–697. 10.1104/pp.103.022988 - DOI - PMC - PubMed
-
- Brautigam A., Shrestha R. P., Whitten D., Wilkerson C. G., Carr K. M., Froehlich J. E., et al. (2008). Low-coverage massively parallel pyrosequencing of cDNAs enables proteomics in non-model species: comparison of a species-specific database generated by pyrosequencing with databases from related species for proteome analysis of pea chloroplast envelopes. J. Biotechnol. 136 44–53. 10.1016/j.jbiotec.2008.02.007 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

