Reconstruction of Oryza sativa indica Genome Scale Metabolic Model and Its Responses to Varying RuBisCO Activity, Light Intensity, and Enzymatic Cost Conditions
- PMID: 29250098
- PMCID: PMC5715477
- DOI: 10.3389/fpls.2017.02060
Reconstruction of Oryza sativa indica Genome Scale Metabolic Model and Its Responses to Varying RuBisCO Activity, Light Intensity, and Enzymatic Cost Conditions
Abstract
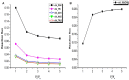
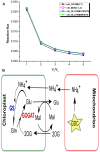
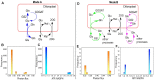
To combat decrease in rice productivity under different stresses, an understanding of rice metabolism is needed. Though there are different genome scale metabolic models (GSMs) of Oryza sativa japonica, no GSM with gene-protein-reaction association exist for Oryza sativa indica. Here, we report a GSM, OSI1136 of O.s. indica, which includes 3602 genes and 1136 metabolic reactions and transporters distributed across the cytosol, mitochondrion, peroxisome, and chloroplast compartments. Flux balance analysis of the model showed that for varying RuBisCO activity (Vc/Vo) (i) the activity of the chloroplastic malate valve increases to transport reducing equivalents out of the chloroplast under increased photorespiratory conditions and (ii) glyceraldehyde-3-phosphate dehydrogenase and phosphoglycerate kinase can act as source of cytosolic ATP under decreased photorespiration. Under increasing light conditions we observed metabolic flexibility, involving photorespiration, chloroplastic triose phosphate and the dicarboxylate transporters of the chloroplast and mitochondrion for redox and ATP exchanges across the intracellular compartments. Simulations under different enzymatic cost conditions revealed (i) participation of peroxisomal glutathione-ascorbate cycle in photorespiratory H2O2 metabolism (ii) different modes of the chloroplastic triose phosphate transporters and malate valve, and (iii) two possible modes of chloroplastic Glu-Gln transporter which were related with the activity of chloroplastic and cytosolic isoforms of glutamine synthetase. Altogether, our results provide new insights into plant metabolism.
Keywords: light intensity; Oryza sativa indica; enzymatic cost; flux balance analysis; metabolic model; photorespiration.
Figures





References
-
- Backhausen J. E., Emmerlich A., Holtgrefe S., Horton P., Müller-Röber B., Scheibe R. (1998). Isolated intact chloroplasts and whole leaves from transgenic potato plants with altered levels of chloroplast NADP-MDH: Interactions between photosynthetic electron transport and malate metabolism. Planta 207 105–114. 10.1007/s004250050461 - DOI
-
- Backhausen J. E., Scheibe R. (1999). Adaptation of tobacco plants to elevated CO2: influence of leaf age on changes in physiology, redox states and NADP-malate dehydrogenase activity. J. Exp. Bot. 50 665–675. 10.1093/jxb/50.334.665 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

