Pharmacological perturbation of CDK9 using selective CDK9 inhibition or degradation
- PMID: 29251720
- PMCID: PMC5912898
- DOI: 10.1038/nchembio.2538
Pharmacological perturbation of CDK9 using selective CDK9 inhibition or degradation
Abstract
Cyclin-dependent kinase 9 (CDK9), an important regulator of transcriptional elongation, is a promising target for cancer therapy, particularly for cancers driven by transcriptional dysregulation. We characterized NVP-2, a selective ATP-competitive CDK9 inhibitor, and THAL-SNS-032, a selective CDK9 degrader consisting of a CDK-binding SNS-032 ligand linked to a thalidomide derivative that binds the E3 ubiquitin ligase Cereblon (CRBN). To our surprise, THAL-SNS-032 induced rapid degradation of CDK9 without affecting the levels of other SNS-032 targets. Moreover, the transcriptional changes elicited by THAL-SNS-032 were more like those caused by NVP-2 than those induced by SNS-032. Notably, compound washout did not significantly reduce levels of THAL-SNS-032-induced apoptosis, suggesting that CDK9 degradation had prolonged cytotoxic effects compared with CDK9 inhibition. Thus, our findings suggest that thalidomide conjugation represents a promising strategy for converting multi-targeted inhibitors into selective degraders and reveal that kinase degradation can induce distinct pharmacological effects compared with inhibition.
Conflict of interest statement
Figures
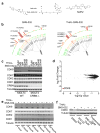
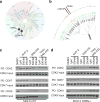
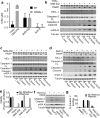
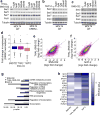
Similar articles
-
The Molecular Context of Vulnerability for CDK9 Suppression in Triple Wild-Type Melanoma.J Invest Dermatol. 2021 Aug;141(8):2018-2027.e4. doi: 10.1016/j.jid.2020.12.035. Epub 2021 Mar 18. J Invest Dermatol. 2021. PMID: 33745909 Free PMC article.
-
Discovery of Potent and Selective CDK9 Degraders for Targeting Transcription Regulation in Triple-Negative Breast Cancer.J Med Chem. 2021 Oct 14;64(19):14822-14847. doi: 10.1021/acs.jmedchem.1c01350. Epub 2021 Sep 20. J Med Chem. 2021. PMID: 34538051
-
Discovery of novel CDK9 inhibitor with tridentate ligand: Design, synthesis and biological evaluation.Bioorg Chem. 2024 Sep;150:107550. doi: 10.1016/j.bioorg.2024.107550. Epub 2024 Jun 10. Bioorg Chem. 2024. PMID: 38878756
-
Recent Developments in the Biology and Medicinal Chemistry of CDK9 Inhibitors: An Update.J Med Chem. 2020 Nov 25;63(22):13228-13257. doi: 10.1021/acs.jmedchem.0c00744. Epub 2020 Sep 17. J Med Chem. 2020. PMID: 32866383 Review.
-
CDK9 a potential target for drug development.Med Chem. 2008 May;4(3):210-8. doi: 10.2174/157340608784325205. Med Chem. 2008. PMID: 18473913 Review.
Cited by
-
Inhibitors of Cyclin-Dependent Kinases: Types and Their Mechanism of Action.Int J Mol Sci. 2021 Mar 10;22(6):2806. doi: 10.3390/ijms22062806. Int J Mol Sci. 2021. PMID: 33802080 Free PMC article. Review.
-
Human promoter directionality is determined by transcriptional initiation and the opposing activities of INTS11 and CDK9.Elife. 2024 Jul 8;13:RP92764. doi: 10.7554/eLife.92764. Elife. 2024. PMID: 38976490 Free PMC article.
-
Synthetic Lethal Interaction of SHOC2 Depletion with MEK Inhibition in RAS-Driven Cancers.Cell Rep. 2019 Oct 1;29(1):118-134.e8. doi: 10.1016/j.celrep.2019.08.090. Cell Rep. 2019. PMID: 31577942 Free PMC article.
-
Evolution of Small Molecule Kinase Drugs.ACS Med Chem Lett. 2018 Dec 18;10(2):153-160. doi: 10.1021/acsmedchemlett.8b00445. eCollection 2019 Feb 14. ACS Med Chem Lett. 2018. PMID: 30783496 Free PMC article.
-
Toward Development of the Male Pill: A Decade of Potential Non-hormonal Contraceptive Targets.Front Cell Dev Biol. 2020 Feb 26;8:61. doi: 10.3389/fcell.2020.00061. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 32161754 Free PMC article. Review.
References
-
- Dorée M. & Galas, S. The cyclin-dependent protein kinases and the control of cell division. The FASEB Journal. 1994;8:1114–1121. - PubMed
-
- Sun T, Co NN, Wong N. PFTK1 interacts with cyclin Y to activate non-canonical Wnt signaling in hepatocellular carcinoma. Biochemical and Biophysical Research Communications. 2014;449:163–168. - PubMed
-
- Liu Y, Cheng K, Gong K, Fu AKY, Ip NY. Pctaire1 Phosphorylates N-Ethylmaleimide-sensitive Fusion Protein: IMPLICATIONS IN THE REGULATION OF ITS HEXAMERIZATION AND EXOCYTOSIS. Journal of Biological Chemistry. 2006;281:9852–9858. - PubMed
-
- Peng J, Marshall NF, Price DH. Identification of a Cyclin Subunit Required for the Function ofDrosophila P-TEFb. Journal of Biological Chemistry. 1998;273:13855–13860. - PubMed
Publication types
MeSH terms
Substances
Associated data
- PubChem-Substance/348356266
- PubChem-Substance/348356277
- PubChem-Substance/348356281
- PubChem-Substance/348356282
- PubChem-Substance/348356283
- PubChem-Substance/348356284
- PubChem-Substance/348356285
- PubChem-Substance/348356286
- PubChem-Substance/348356287
- PubChem-Substance/348356267
- PubChem-Substance/348356268
- PubChem-Substance/348356269
- PubChem-Substance/348356270
- PubChem-Substance/348356271
- PubChem-Substance/348356272
- PubChem-Substance/348356273
- PubChem-Substance/348356274
- PubChem-Substance/348356275
- PubChem-Substance/348356276
- PubChem-Substance/348356278
- PubChem-Substance/348356279
- PubChem-Substance/348356280
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

