Informatics for cancer immunotherapy
- PMID: 29253114
- PMCID: PMC5834022
- DOI: 10.1093/annonc/mdx682
Informatics for cancer immunotherapy
Abstract
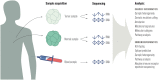
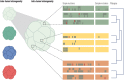
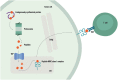
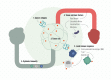
The rapid development of immunomodulatory cancer therapies has led to a concurrent increase in the application of informatics techniques to the analysis of tumors, the tumor microenvironment, and measures of systemic immunity. In this review, the use of tumors to gather genetic and expression data will first be explored. Next, techniques to assess tumor immunity are reviewed, including HLA status, predicted neoantigens, immune microenvironment deconvolution, and T-cell receptor sequencing. Attempts to integrate these data are in early stages of development and are discussed in this review. Finally, we review the application of these informatics strategies to therapy development, with a focus on vaccines, adoptive cell transfer, and checkpoint blockade therapies.
Keywords: adoptive cell transfer; bioinformatics; checkpoint blockade; computational biology; immunotherapy; neoantigens.
© The Author 2017. Published by Oxford University Press on behalf of the European Society for Medical Oncology. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures

Similar articles
-
Prospects for personalized combination immunotherapy for solid tumors based on adoptive cell therapies and immune checkpoint blockade therapies.Nihon Rinsho Meneki Gakkai Kaishi. 2017;40(1):68-77. doi: 10.2177/jsci.40.68. Nihon Rinsho Meneki Gakkai Kaishi. 2017. PMID: 28539557 Review.
-
New emerging targets in cancer immunotherapy: the role of neoantigens.ESMO Open. 2020 Apr;4(Suppl 3):e000684. doi: 10.1136/esmoopen-2020-000684. ESMO Open. 2020. PMID: 32269031 Free PMC article. Review.
-
Tumor neoantigens: Novel strategies for application of cancer immunotherapy.Oncol Res. 2023 Jun 27;31(4):437-448. doi: 10.32604/or.2023.029924. eCollection 2023. Oncol Res. 2023. PMID: 37415744 Free PMC article. Review.
-
Integrating Bioinformatics Strategies in Cancer Immunotherapy: Current and Future Perspectives.Comb Chem High Throughput Screen. 2020;23(8):687-698. doi: 10.2174/1386207323666200427113734. Comb Chem High Throughput Screen. 2020. PMID: 32338212 Review.
-
Targeting Neoepitopes to Treat Solid Malignancies: Immunosurgery.Front Immunol. 2021 Jul 15;12:592031. doi: 10.3389/fimmu.2021.592031. eCollection 2021. Front Immunol. 2021. PMID: 34335558 Free PMC article. Review.
Cited by
-
Genetic engineering cellular vesicles expressing CD64 as checkpoint antibody carrier for cancer immunotherapy.Theranostics. 2021 Apr 7;11(12):6033-6043. doi: 10.7150/thno.48868. eCollection 2021. Theranostics. 2021. PMID: 33897897 Free PMC article.
-
Tumor-intrinsic SIRPA promotes sensitivity to checkpoint inhibition immunotherapy in melanoma.Cancer Cell. 2022 Nov 14;40(11):1324-1340.e8. doi: 10.1016/j.ccell.2022.10.012. Epub 2022 Nov 3. Cancer Cell. 2022. PMID: 36332624 Free PMC article.
-
Comprehensive co-expression analysis reveals TMC8 as a prognostic immune-associated gene in head and neck squamous cancer.Oncol Lett. 2021 Jul;22(1):498. doi: 10.3892/ol.2021.12759. Epub 2021 Apr 27. Oncol Lett. 2021. PMID: 33981360 Free PMC article.
-
Advances in Therapeutic Cancer Vaccines, Their Obstacles, and Prospects Toward Tumor Immunotherapy.Mol Biotechnol. 2025 Apr;67(4):1336-1366. doi: 10.1007/s12033-024-01144-3. Epub 2024 Apr 16. Mol Biotechnol. 2025. PMID: 38625508 Review.
-
KLF12 overcomes anti-PD-1 resistance by reducing galectin-1 in cancer cells.J Immunother Cancer. 2023 Aug;11(8):e007286. doi: 10.1136/jitc-2023-007286. J Immunother Cancer. 2023. PMID: 37586772 Free PMC article.
References
-
- Stratton MR, Campbell PJ, Futreal PA.. The cancer genome. Nature 2009; 458(7239): 719–724.http://dx.doi.org/10.1038/nature07943 - DOI - PMC - PubMed
-
- Mardis ER. Genome sequencing and cancer. Curr Opin Genet Dev 2012; 22(3): 245–250.http://dx.doi.org/10.1016/j.gde.2012.03.005 - DOI - PMC - PubMed
-
- Ding L, Wendl MC, McMichael JF, Raphael BJ.. Expanding the computational toolbox for mining cancer genomes. Nat Rev Genet 2014; 15(8): 556–570.http://dx.doi.org/10.1038/nrg3767 - DOI - PMC - PubMed
-
- Martincorena I, Roshan A, Gerstung M. et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 2015; 348(6237): 880–886.http://dx.doi.org/10.1126/science.aaa6806 - DOI - PMC - PubMed
-
- Carter SL, Cibulskis K, Helman E. et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol 2012; 30(5): 413–421.http://dx.doi.org/10.1038/nbt.2203 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

