FANCD2 binding identifies conserved fragile sites at large transcribed genes in avian cells
- PMID: 29253234
- PMCID: PMC5815096
- DOI: 10.1093/nar/gkx1260
FANCD2 binding identifies conserved fragile sites at large transcribed genes in avian cells
Abstract
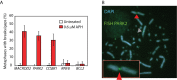
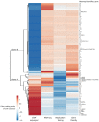
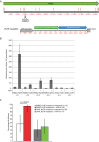
Common Chromosomal Fragile Sites (CFSs) are specific genomic regions prone to form breaks on metaphase chromosomes in response to replication stress. Moreover, CFSs are mutational hotspots in cancer genomes, showing that the mutational mechanisms that operate at CFSs are highly active in cancer cells. Orthologs of human CFSs are found in a number of other mammals, but the extent of CFS conservation beyond the mammalian lineage is unclear. Characterization of CFSs from distantly related organisms can provide new insight into the biology underlying CFSs. Here, we have mapped CFSs in an avian cell line. We find that, overall the most significant CFSs coincide with extremely large conserved genes, from which very long transcripts are produced. However, no significant correlation between any sequence characteristics and CFSs is found. Moreover, we identified putative early replicating fragile sites (ERFSs), which is a distinct class of fragile sites and we developed a fluctuation analysis revealing high mutation rates at the CFS gene PARK2, with deletions as the most prevalent mutation. Finally, we show that avian homologs of the human CFS genes despite their fragility have resisted the general intron size reduction observed in birds suggesting that CFSs have a conserved biological function.
Figures



References
-
- Minocherhomji S., Hickson I.D.. Structure-specific endonucleases: guardians of fragile site stability. Trends Cell Biol. 2014; 24:321–327. - PubMed
-
- Debatisse M., Le Tallec B., Letessier A., Dutrillaux B., Brison O.. Common fragile sites: mechanisms of instability revisited. Trends Genet. 2012; 28:22–32. - PubMed
-
- Torres-Rosell J., De Piccoli G., Cordon-Preciado V., Farmer S., Jarmuz A., Machin F., Pasero P., Lisby M., Haber J.E., Aragon L.. Anaphase onset before complete DNA replication with intact checkpoint responses. Science. 2007; 315:1411–1415. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

