Mitochondrial CDP-diacylglycerol synthase activity is due to the peripheral protein, TAMM41 and not due to the integral membrane protein, CDP-diacylglycerol synthase 1
- PMID: 29253589
- PMCID: PMC5791848
- DOI: 10.1016/j.bbalip.2017.12.005
Mitochondrial CDP-diacylglycerol synthase activity is due to the peripheral protein, TAMM41 and not due to the integral membrane protein, CDP-diacylglycerol synthase 1
Abstract
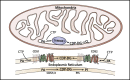
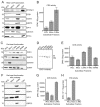
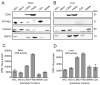
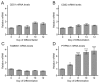
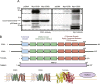
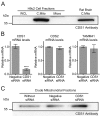
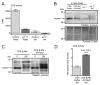
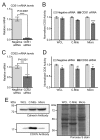
CDP diacylglycerol synthase (CDS) catalyses the conversion of phosphatidic acid (PA) to CDP-diacylglycerol, an essential intermediate in the synthesis of phosphatidylglycerol, cardiolipin and phosphatidylinositol (PI). CDS activity has been identified in mitochondria and endoplasmic reticulum of mammalian cells apparently encoded by two highly-related genes, CDS1 and CDS2. Cardiolipin is exclusively synthesised in mitochondria and recent studies in cardiomyocytes suggest that the peroxisome proliferator-activated receptor γ coactivator 1 (PGC-1α and β) serve as transcriptional regulators of mitochondrial biogenesis and up-regulate the transcription of the CDS1 gene. Here we have examined whether CDS1 is responsible for the mitochondrial CDS activity. We report that differentiation of H9c2 cells with retinoic acid towards cardiomyocytes is accompanied by increased expression of mitochondrial proteins, oxygen consumption, and expression of the PA/PI binding protein, PITPNC1, and CDS1 immunoreactivity. Both CDS1 immunoreactivity and CDS activity were found in mitochondria of H9c2 cells as well as in rat heart, liver and brain mitochondria. However, the CDS1 immunoreactivity was traced to a peripheral p55 cross-reactive mitochondrial protein and the mitochondrial CDS activity was due to a peripheral mitochondrial protein, TAMM41, not an integral membrane protein as expected for CDS1. TAMM41 is the mammalian equivalent of the recently identified yeast protein, Tam41. Knockdown of TAMM41 resulted in decreased mitochondrial CDS activity, decreased cardiolipin levels and a decrease in oxygen consumption. We conclude that the CDS activity present in mitochondria is mainly due to TAMM41, which is required for normal mitochondrial function.
Keywords: Cardiolipin; Differentiation; Heart; Mitochondria; PITPNC1; Phosphatidic acid; Retinoic acid.
Copyright © 2017 The Author(s). Published by Elsevier B.V. All rights reserved.
Figures



References
-
- Mejia E.M., Nguyen H., Hatch G.M. Mammalian cardiolipin biosynthesis. Chem. Phys. Lipids. 2014;179:11–16. - PubMed
-
- Palmer J.W., Tandler B., Hoppel C.L. Biochemical properties of subsarcolemmal and interfibrillar mitochondria isolated from rat cardiac muscle. J. Biol. Chem. 1977;252:8731–8739. - PubMed
-
- Hock M.B., Kralli A. Transcriptional control of mitochondrial biogenesis and function. Annu. Rev. Physiol. 2009;71:177–203. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

