Fundamental properties of the mammalian innate immune system revealed by multispecies comparison of type I interferon responses
- PMID: 29253856
- PMCID: PMC5747502
- DOI: 10.1371/journal.pbio.2004086
Fundamental properties of the mammalian innate immune system revealed by multispecies comparison of type I interferon responses
Abstract
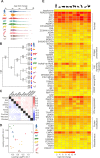
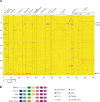
The host innate immune response mediated by type I interferon (IFN) and the resulting up-regulation of hundreds of interferon-stimulated genes (ISGs) provide an immediate barrier to virus infection. Studies of the type I 'interferome' have mainly been carried out at a single species level, often lacking the power necessary to understand key evolutionary features of this pathway. Here, using a single experimental platform, we determined the properties of the interferomes of multiple vertebrate species and developed a webserver to mine the dataset. This approach revealed a conserved 'core' of 62 ISGs, including genes not previously associated with IFN, underscoring the ancestral functions associated with this antiviral host response. We show that gene expansion contributes to the evolution of the IFN system and that interferomes are shaped by lineage-specific pressures. Consequently, each mammal possesses a unique repertoire of ISGs, including genes common to all mammals and others unique to their specific species or phylogenetic lineages. An analysis of genes commonly down-regulated by IFN suggests that epigenetic regulation of transcription is a fundamental aspect of the IFN response. Our study provides a resource for the scientific community highlighting key paradigms of the type I IFN response.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Woolhouse MEJ, Gowtage-Sequeria S. Host range and emerging and reemerging pathogens. Emerg Infect Dis. Centers for Disease Control and Prevention; 2005;11: 1842–7. doi: 10.3201/eid1112.050997 - DOI - PMC - PubMed
-
- Reperant LA, Brown IH, Haenen OL, de Jong MD, Osterhaus ADME, Papa A, et al. Companion Animals as a Source of Viruses for Human Beings and Food Production Animals. J Comp Pathol. 2016;155: S41–S53. doi: 10.1016/j.jcpa.2016.07.006 - DOI - PubMed
-
- Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. NIH Public Access; 2011;472: 481–5. doi: 10.1038/nature09907 - DOI - PMC - PubMed
-
- Schoggins JW, Rice CM. Interferon-stimulated genes and their antiviral effector functions. Curr Opin Virol. NIH Public Access; 2011;1: 519–25. doi: 10.1016/j.coviro.2011.10.008 - DOI - PMC - PubMed
-
- Daugherty MD, Malik HS. Rules of Engagement: Molecular Insights from Host-Virus Arms Races. Annu Rev Genet. Annual Reviews; 2012;46: 677–700. doi: 10.1146/annurev-genet-110711-155522 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

