The ubiquitous 'cancer mutational signature' 5 occurs specifically in cancers with deleted FHIT alleles
- PMID: 29254236
- PMCID: PMC5731946
- DOI: 10.18632/oncotarget.22321
The ubiquitous 'cancer mutational signature' 5 occurs specifically in cancers with deleted FHIT alleles
Abstract
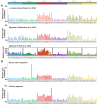
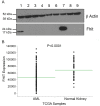
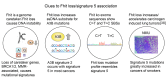
The FHIT gene is located at the fragile FRA3B locus where activation by carcinogen-induced and endogenous replication stress causes FHIT deletions even in normal cells over a lifetime. Our lab has shown that loss of FHIT expression causes genome instability and provides single-strand DNA substrates for APOBEC3B hypermutation, in line with evidence that FHIT locus deletions occur in many cancers. Based on these biological features, we hypothesized that FHIT loss drives development of COSMIC mutational signature 5 and here provide evidence, including data mining of >6,500 TCGA samples, that FHIT is the cancer-associated gene with copy number alterations correlating most significantly with signature 5 mutation rate. In addition, tissues of Fhit-deficient mice exhibit a mutational signature strongly resembling signature 5 (cosine similarity value = 0.89). We conclude that FHIT loss is a molecular determinant for signature 5 mutations, which occur in all cancer types early in cancer development, are clock-like, and accelerated by carcinogen exposure. Loss of FHIT caretaker function may be a predictive and preventive marker for cancer development.
Keywords: cancer genes; clock-like signatures; data mining; exome sequences; mutational signatures.
Conflict of interest statement
CONFLICTS OF INTEREST The authors confirm that there are no conflicts of interest.
Figures

References
-
- Ohta M, Inoue H, Cotticelli MG, Kastury K, Baffa R, Palazzo J, Siprashvili Z, Mori M, McCue P, Druck T, Croce CM, Huebner K. The FHIT gene, spanning the chromosome 3p14.2 fragile site and renal carcinoma-associated t(3;8) breakpoint, is abnormal in digestive tract cancers. Cell. 1996;84:87–97. - PubMed
-
- Sozzi G, Pastorino U, Moiraghi L, Tagliabue E, Pezzella F, Ghirelli C, Tornielli S, Sard L, Huebner K, Pierotti MA, Croce CM, Pilotti S. Loss of FHIT function in lung cancer and preinvasive bronchial lesions. Cancer Res. 1997;58:5032–7. - PubMed
-
- Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA, Jr, Kastrinakis NG, Levy B, Kletsas D, Yoneta A, Herlyn M, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–13. - PubMed
-
- Guler G, Uner A, Guler N, Han SY, Iliopoulos D, McCue P, Huebner K. Concordant loss of fragile gene expression early in breast cancer development. Pathol Int. 2005;55:471–8. - PubMed
-
- Bartkova J, Horejsí Z, Koed K, Krämer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, Ørntoft T, Lukas J, Bartek J. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–70. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

