Landscape of nuclear transport receptor cargo specificity
- PMID: 29254951
- PMCID: PMC5740495
- DOI: 10.15252/msb.20177608
Landscape of nuclear transport receptor cargo specificity
Abstract
Nuclear transport receptors (NTRs) recognize localization signals of cargos to facilitate their passage across the central channel of nuclear pore complexes (NPCs). About 30 different NTRs constitute different transport pathways in humans and bind to a multitude of different cargos. The exact cargo spectrum of the majority of NTRs, their specificity and even the extent to which active nucleocytoplasmic transport contributes to protein localization remains understudied because of the transient nature of these interactions and the wide dynamic range of cargo concentrations. To systematically map cargo-NTR relationships in situ, we used proximity ligation coupled to mass spectrometry (BioID). We systematically fused the engineered biotin ligase BirA* to 16 NTRs. We estimate that a considerable fraction of the human proteome is subject to active nuclear transport. We quantified the specificity and redundancy in NTR interactions and identified transport pathways for cargos. We extended the BioID method by the direct identification of biotinylation sites. This approach enabled us to identify interaction interfaces and to discriminate direct versus piggyback transport mechanisms. Data are available via ProteomeXchange with identifier PXD007976.
Keywords: interaction network; nuclear pore complex; protein transport; proteomics; proximity ligation.
© 2017 European Molecular Biology Laboratory. Published under the terms of the CC BY 4.0 license.
Figures

Scheme showing the composition and approximate position of nucleoporin subcomplexes with respect to the overall structure (Protein Data Bank EMD‐3103). FG‐Nups are underlined.
Overview of the nucleocytoplasmic transport pathways and NTRs. Cargos are either bound by importin βs and exportins or bound by importin αs that serve as an adaptor for KPNB1. Recycling of RAN‐GDP by NTF2 into the nucleus and importin αs by XPO2 back to the cytoplasm is shown.

Scheme of the general workflow used in this study showing the molecular cloning, proximity ligation in situ and affinity capture of biotinylated proteins on streptavidin sepharose beads. Isolated proteins were on‐bead digested using trypsin for indirect identification and biotinylated peptides eluted in an additional step using a mixture of ACN and TFA for direct identification, both by MS.
Stable cell lines expressing transport factors fused to BirA* were fixed and stained with Streptavidin‐Alexa 647 (red) to visualize the subcellular localization of biotinylated proteins, the overexpressed BirA* fusion proteins (anti‐FLAG, green), and the DNA (Hoechst, blue). Scale bar, 10 μm.
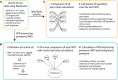
Seventeen different transport factors and in total 28 N‐ and C‐terminal BirA* fusion proteins and four control samples were measured in biological quadruplicate and technical duplicate. Protein intensities calculated using MaxQuant were used for further analysis.
The NTR‐interacting proteome (NIP) and background proteome were calculated by quantification of NTR samples against the control set. The NIP does cover all NTR‐interacting proteins regardless of how many NTRs they interact with.
Procedure to calculate protein specificity scores for each NTR. Proteins that interact with multiple NTRs are penalized (see Materials and Methods for details). The workflow for the direct identification of biotinylated peptides is not depicted in the figure.

- A, B
GO enrichment analysis for the terms “cellular component” (A) and “molecular function” (B). The top 10 most significant GO terms are plotted for the NIP or background proteome. Numbers in (B) refer to (1) the aldehyde or oxo group of donors, (2) the aldehyde or oxo group of donors, NAD or NADP as acceptor, (3) NAD(P)H. The NIP is significantly enriched for nuclear proteins.
- C
RNC score from Wühr et al (cytoplasmic to nuclear from 0 to 1) compared to all significantly identified proteins in the NIP and background proteome. The NIP has a trend toward high RNC values indicating a high content of proteins that show preferential nuclear localization, while to opposite is the case for the background proteome.
- D
MW distribution of all proteins in the NIP (median = 74 kDa) and background (median = 54 kDa) compared to all the reviewed human proteins in the UniProt database (median = 46 kDa). The NIP shows a trend toward higher MW even though protein complex association (so‐called native MW) is not considered in this plot.
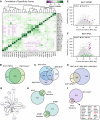
- A–D
(A) Pearson correlation of the specificity scores of all 28 experiments (excluding the four controls). N‐ and C‐terminally tagged cell lines and functionally related NTRs show a high degree of similarity. The overlap of significant identifications of N‐ and C‐terminally tagged TNPO2 (B) between different importin αs (C) and KPNB1 with importin αs (D) is shown as representative examples. Importin αs show a high degree functional redundancy, and a prominent fraction of significant identifications for KPNB1 overlap with hits from importin αs that are adaptors for cargo binding for KPNB1.
- E, F
iBAQ and specificity scores in BirA*‐KPNB1 and BirA*‐IPO5 for selected protein complexes or group of proteins are shown as scatter plots. Common interaction partners like Nups get penalized by specificity score calculation because they interact with multiple NTRs.
- G
Interactions of XPO2 with other NTRs recover known properties of the nucleocytoplasmic transport system, including interactions with importin αs. Arrow thickness is proportional to the specificity score of the interaction. Arrow direction indicates bait (source)–prey (target) relationships. Two arrows pointing in the same direction indicate the N‐ or C‐terminally tagged version of the NTR retrieving the same prey.
- H, I
Comparison of cargos known from literature for IPO5, KPNB1 (Kimura et al, 2017), and XPO1 (Thakar et al, 2013; Kırlı et al, 2015; Wühr et al, 2015). For XPO1, only cargos significant in at least two out of the three previous large‐scale studies were considered. Proteins highlighted in red are well‐established XPO1 cargos.
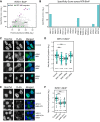
iBAQ and specificity scores identified with INTS11‐BirA* are shown as scatter plot; members of the integrator complex and NTRs are highlighted.
Specificity scores obtained with various NTR‐BirA* fusion proteins for INTS11.
Subcellular distribution of the INTS11‐BirA* upon siRNA treatment against IMA1, IMA5, IPO5, and a negative control (scrambled siRNA). Importin αs induce a shift of INTS11 toward the cytoplasm.
Quantification of the ratio of nucleoplasmic to cytoplasmic (N/C) distribution of INTS11 upon siRNA treatment (**Wilcoxon signed‐rank test P‐value < 0.01).
Subcellular distribution of INTS11‐BirA* upon removal and replacement of the predicted cNLS with a linker. The absence of the predicted cNLS leads to an almost exclusive cytoplasmic localization of INTS11.
Quantification of the N/C ratio upon cNLS removal (**Wilcoxon signed‐rank test P‐value < 0.01).
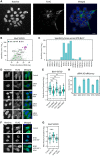
Automated detection of cells using the Hoechst staining as reference by CellProfiler. The overlaid numbers are the mean intensity in the cytoplasmic and nuclear area of anti‐FLAG, which were used to exclude non‐expressing cells.
iBAQ and specificity scores obtained with BirA*‐EIF3D as scatter plot; members of eukaryotic translation initiation factor 3 and NTRs are highlighted. Importin βs are significantly enriched for EIF3D.
Specificity score for EIF3D obtained with various NTR‐BirA* fusion proteins. IPO7 and IPO8 identified in the reciprocal BirA* AP were not part of the initial data set.
Subcellular distribution of the BirA*‐EIF3D upon siRNA treatment against IPO4, IPO5, IPO11, TNPO2, and a negative control (scrambled siRNA). IPO5 and IPO11 induce the strongest shift of EIF3D toward the cytoplasm.
Quantification of the ratio of nucleoplasmic to cytoplasmic (N/C) distribution of EIF3D upon siRNA treatment (**Wilcoxon signed‐rank test P‐value < 0.01).
Subcellular distribution of BirA*‐EIF3D upon removal and replacement of a DE‐rich motif predicted by DILIMOT with a linker. The deletion but not substitution of the predicted DE‐rich motif leads to a nucleoplasmic retention.
Quantification of the N/C ratio upon DE‐rich motif removal (**Wilcoxon signed‐rank test P‐value < 0.01).
Bar chart of knock‐down efficiency after siRNA mediated depletion of selected NTRs (IMA1, IMA5, IPO11, IPO4, IPO5, and TNPO2). Transcript levels were quantified by qPCR, and they are expressed as knock‐down efficiency. The knock‐down efficiency ranges from 39% (IMA5) to 94% (IMA1) after 3 days of treatment. Error bars indicate standard deviation. N = 3.
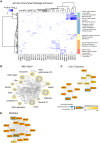
- A
Comparison of GO terms enriched among proteins interacting specifically with NTRs. GO enrichment was performed individually for each NTR sample by ranking proteins according to their specificity scores. Significant GO terms (P‐value < 0.001) from the category “biological process” were combined and compared across NTRs. Distinct biological processes display specific association with related NTRs.
- B
Network analysis of the top 2% enriched proteins of the IMA1‐BirA* experiment. Various import cargos are found associated with IMA1. LSm (like Sm), BAF (BRG1‐ or HBRM‐associated factors), NuRD (nucleosome remodeling deacetylase), TF (transcription factor), TFIID (transcription factor II D).
- C, D
Selected subnetworks are highlighted; specificity scores are indicated by color gradients and detected biotinylated proteins by blue frames.

- A–E
Network analysis of the top 2% enriched proteins for IMA5‐BirA* (A), BirA*‐IPO5 (B), IPO4‐BirA* (C), BirA*‐KPNB1 (D), and NTF2‐BirA* (E). Selected subnetworks are shown with the specificity score (color gradient) and detected biotinylated peptides (blue frame). MCM (minichromosome maintenance), ORC (origin recognition complex), eIF3 (eukaryotic translation initiation factor 3), APC (anaphase‐promoting complex), Arf GAP (ADP‐ribosylation factor GTPase‐activating proteins), WAVE (WASP‐family verprolin homologous protein).

- A–C
Same as Fig EV3 but for XPO1‐BirA* (A), XPO2‐BirA (B) and BirA*‐XPO7 (C). PCAF (p300/CBP‐associated factor), AMPK (AMP‐activated protein kinase), COP9 (constitutive photomorphogenesis 9).

Specificity scores of selected protein complexes and functionally related proteins are shown as a heat map across all experiments. Multiple subunits of the depicted protein complexes are identified to interact with specific NTRs [TCP (=CCT chaperonin containing TCP‐1), ArfGAP (ADP‐ribosylation factor GTPase‐activating proteins), COP9 (constitutive photomorphogenesis 9), CREB (cAMP response element‐binding protein), eIF3 (eukaryotic initiation factor 3)]. Immunofluorescence staining supports preferential biotinylation of nucleolar proteins by certain NTRs (Fig 2B).
Cluster analysis of transcription factors (TF) significantly enriched (adj. Fisher P‐value < 0.01) in at least two experiments are shown. Selected clusters and related TFs therein are highlighted. Related TFs use the same or similar NTR.

- A
NTRs interact with specific structures of the NPC. Protein abundances (iBAQ scores) of Nups across NTR samples often correlate with the number of identified biotinylated peptides.
- B, C
Normalized and median centered intensities of biotinylated peptides of NUP50 and NUP153. Structural domains like the importin α binding site (BD1 and BD2), RAN‐binding domain, zinc fingers, and FG‐repeats are indicated. IPO4, IPO5, and IPO11 show less pronounced intensities as compared to Nup358 and Nup214 (Fig EV3).

- A–C
Same as Fig 8 but for Nup214 (A), Nup98 (B), and Nup358 (C). Structural domains like seven bladed propeller, leucine zippers, autoproteolytic domain, tetratricopeptide repeats (TPR), and Ran‐binding domains (RanBD) are highlighted as well as FG‐repeats.
References
-
- Beck M, Hurt E (2017) The nuclear pore complex: understanding its function through structural insight. Nat Rev Mol Cell Biol 18: 73–89 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

