GNA13 expression promotes drug resistance and tumor-initiating phenotypes in squamous cell cancers
- PMID: 29255247
- PMCID: PMC6168473
- DOI: 10.1038/s41388-017-0038-6
GNA13 expression promotes drug resistance and tumor-initiating phenotypes in squamous cell cancers
Abstract
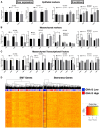
Treatment failure in solid tumors occurs due to the survival of specific subpopulations of cells that possess tumor-initiating (TIC) phenotypes. Studies have implicated G protein-coupled-receptors (GPCRs) in cancer progression and the acquisition of TIC phenotypes. Many of the implicated GPCRs signal through the G protein GNA13. In this study, we demonstrate that GNA13 is upregulated in many solid tumors and impacts survival and metastases in patients. GNA13 levels modulate drug resistance and TIC-like phenotypes in patient-derived head and neck squamous cell carcinoma (HNSCC) cells in vitro and in vivo. Blockade of GNA13 expression, or of select downstream pathways, using small-molecule inhibitors abrogates GNA13-induced TIC phenotypes, rendering cells vulnerable to standard-of-care cytotoxic therapies. Taken together, these data indicate that GNA13 expression is a potential prognostic biomarker for tumor progression, and that interfering with GNA13-induced signaling provides a novel strategy to block TICs and drug resistance in HNSCCs.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

