Identification of H3K4me1-associated proteins at mammalian enhancers
- PMID: 29255264
- PMCID: PMC6007000
- DOI: 10.1038/s41588-017-0015-6
Identification of H3K4me1-associated proteins at mammalian enhancers
Abstract
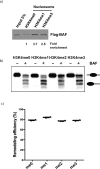
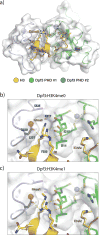
Enhancers act to regulate cell-type-specific gene expression by facilitating the transcription of target genes. In mammalian cells, active or primed enhancers are commonly marked by monomethylation of histone H3 at lysine 4 (H3K4me1) in a cell-type-specific manner. Whether and how this histone modification regulates enhancer-dependent transcription programs in mammals is unclear. In this study, we conducted SILAC mass spectrometry experiments with mononucleosomes and identified multiple H3K4me1-associated proteins, including many involved in chromatin remodeling. We demonstrate that H3K4me1 augments association of the chromatin-remodeling complex BAF to enhancers in vivo and that, in vitro, H3K4me1-marked nucleosomes are more efficiently remodeled by the BAF complex. Crystal structures of the BAF component BAF45C indicate that monomethylation, but not trimethylation, is accommodated by BAF45C's H3K4-binding site. Our results suggest that H3K4me1 has an active role at enhancers by facilitating binding of the BAF complex and possibly other chromatin regulators.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




Comment in
-
Is H3K4me1 at enhancers correlative or causative?Nat Genet. 2018 Jan;50(1):4-5. doi: 10.1038/s41588-017-0018-3. Nat Genet. 2018. PMID: 29273804 No abstract available.
References
-
- Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

