Comparison of mitochondrial gene expression and polysome loading in different tobacco tissues
- PMID: 29255478
- PMCID: PMC5729415
- DOI: 10.1186/s13007-017-0257-4
Comparison of mitochondrial gene expression and polysome loading in different tobacco tissues
Abstract
Background: To investigate translational regulation of gene expression in plant mitochondria, a mitochondrial polysome isolation protocol was established for tobacco to investigate polysomal mRNA loading as a proxy for translational activity. Furthermore, we developed an oligonucleotide based microarray platform to determine the level of Nicotiana tabacum and Arabidopsis thaliana mitochondrial mRNA.
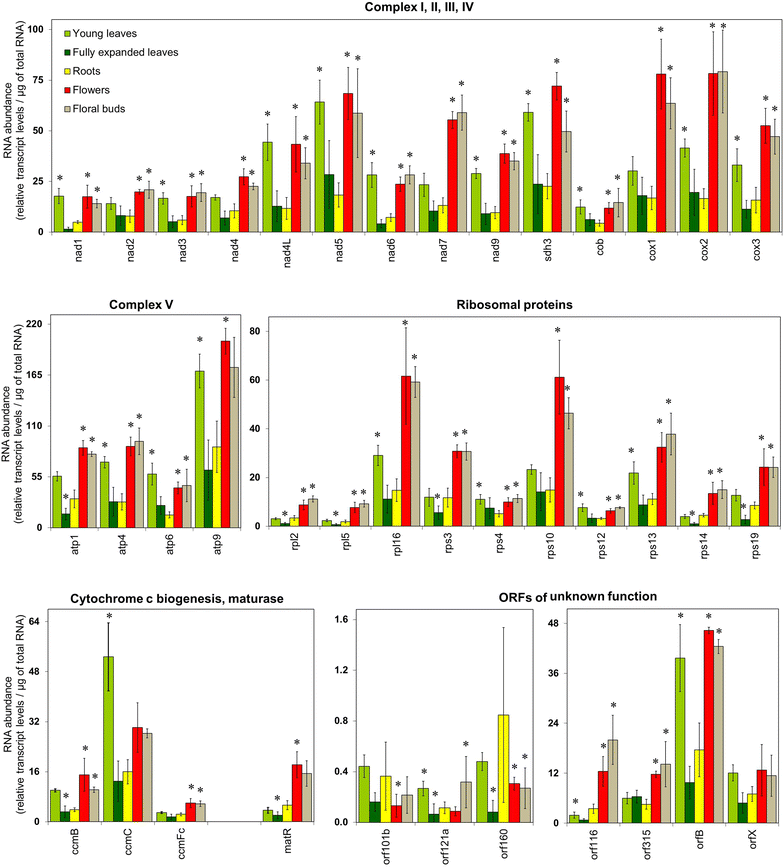
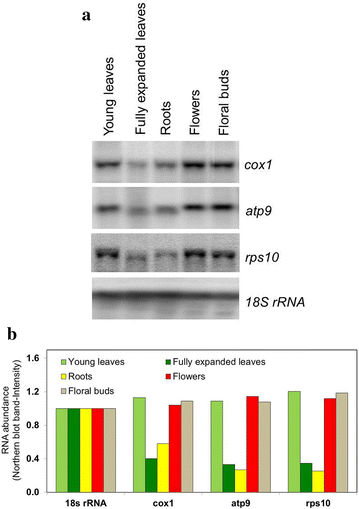
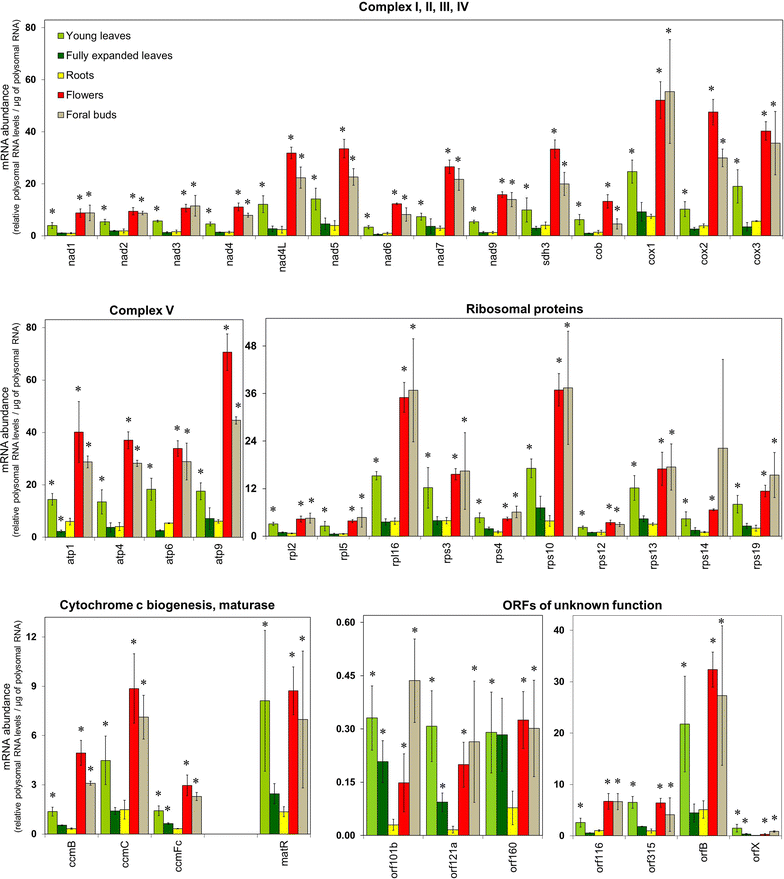
Results: Microarray analysis of free and polysomal mRNAs was used to characterize differences in the levels of free transcripts and ribosome-bound mRNAs in various organs of tobacco plants. We have observed higher mitochondrial transcript levels in young leaves, flowers and floral buds as compared to fully expanded leaves and roots. A similar pattern of abundance was observed for ribosome-bound mitochondrial mRNAs in these tissues. However, the accumulation of the mitochondrial protein COX2 was found to be inversely related to that of its ribosome-bound mRNA.
Conclusions: Our results indicate that the association of mitochondrial mRNAs to ribosomes is largely determined by the total transcript level of a gene. However, at least for Cox2, we demonstrated that the level of ribosome-bound mRNA is not reflected by the amount of COX2 protein.
Keywords: Mitochondrial microarray; Mitochondrial ribosomes; Plant mitochondria; Transcription; Translation.
Figures




Similar articles
-
mRNA sequence features that contribute to translational regulation in Arabidopsis.Nucleic Acids Res. 2005 Feb 16;33(3):955-65. doi: 10.1093/nar/gki240. Print 2005. Nucleic Acids Res. 2005. PMID: 15716313 Free PMC article.
-
Changes in Polysome Association of mRNA Throughout Growth and Development in Arabidopsis thaliana.Plant Cell Physiol. 2015 Nov;56(11):2169-80. doi: 10.1093/pcp/pcv133. Epub 2015 Sep 26. Plant Cell Physiol. 2015. PMID: 26412777
-
Genome-wide analysis of transcript abundance and translation in Arabidopsis seedlings subjected to oxygen deprivation.Ann Bot. 2005 Sep;96(4):647-60. doi: 10.1093/aob/mci217. Epub 2005 Aug 4. Ann Bot. 2005. PMID: 16081496 Free PMC article.
-
Isolation of plant polysomal mRNA by differential centrifugation and ribosome immunopurification methods.Methods Mol Biol. 2009;553:109-26. doi: 10.1007/978-1-60327-563-7_6. Methods Mol Biol. 2009. PMID: 19588103 Review.
-
Translatome profiling: methods for genome-scale analysis of mRNA translation.Brief Funct Genomics. 2016 Jan;15(1):22-31. doi: 10.1093/bfgp/elu045. Epub 2014 Nov 6. Brief Funct Genomics. 2016. PMID: 25380596 Review.
Cited by
-
Experimental approaches to studying translation in plant semi-autonomous organelles.J Exp Bot. 2024 Sep 11;75(17):5175-5187. doi: 10.1093/jxb/erae151. J Exp Bot. 2024. PMID: 38592734 Free PMC article. Review.
-
Particle bombardment-assisted peptide-mediated gene transfer for highly efficient transient assay.BMC Res Notes. 2023 Apr 6;16(1):46. doi: 10.1186/s13104-023-06320-3. BMC Res Notes. 2023. PMID: 37024962 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

