Comparison of mitochondrial gene expression and polysome loading in different tobacco tissues
- PMID: 29255478
- PMCID: PMC5729415
- DOI: 10.1186/s13007-017-0257-4
Comparison of mitochondrial gene expression and polysome loading in different tobacco tissues
Abstract
Background: To investigate translational regulation of gene expression in plant mitochondria, a mitochondrial polysome isolation protocol was established for tobacco to investigate polysomal mRNA loading as a proxy for translational activity. Furthermore, we developed an oligonucleotide based microarray platform to determine the level of Nicotiana tabacum and Arabidopsis thaliana mitochondrial mRNA.
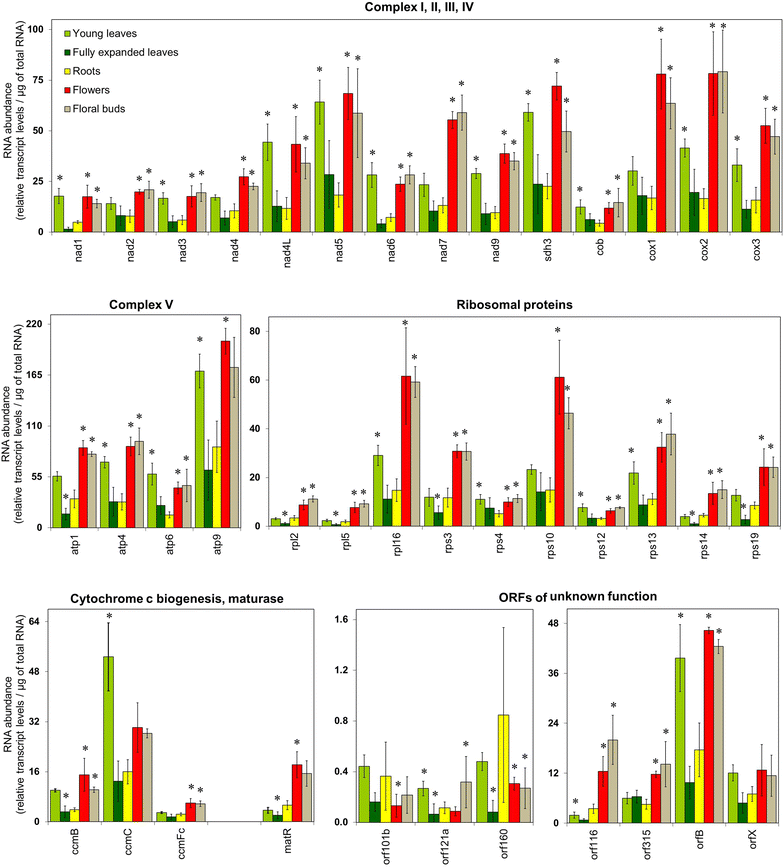
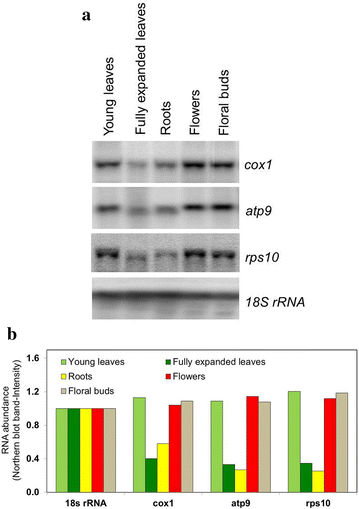
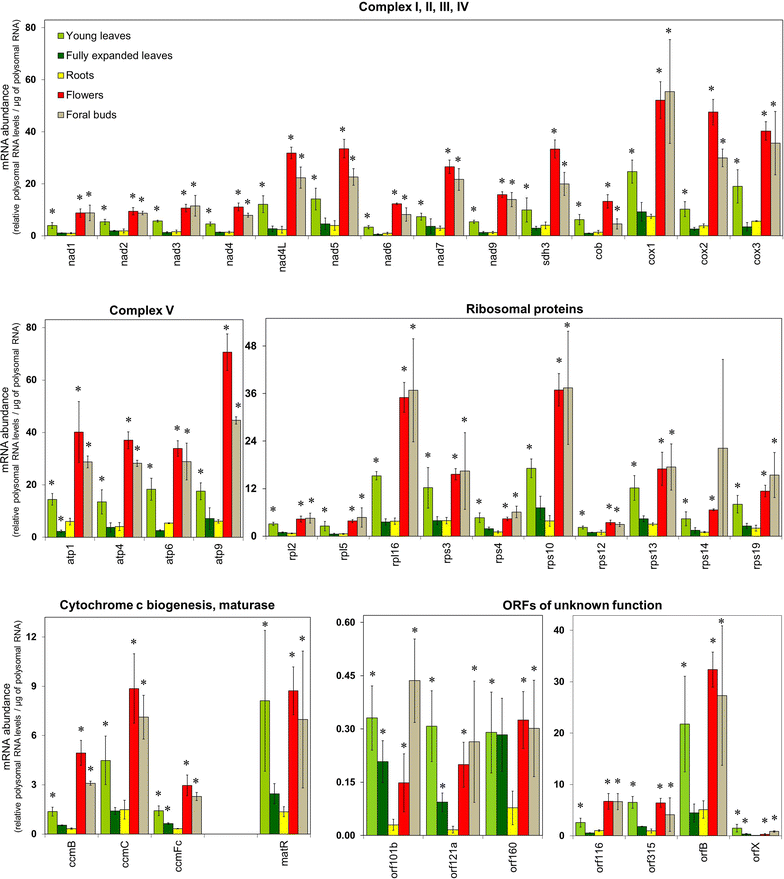
Results: Microarray analysis of free and polysomal mRNAs was used to characterize differences in the levels of free transcripts and ribosome-bound mRNAs in various organs of tobacco plants. We have observed higher mitochondrial transcript levels in young leaves, flowers and floral buds as compared to fully expanded leaves and roots. A similar pattern of abundance was observed for ribosome-bound mitochondrial mRNAs in these tissues. However, the accumulation of the mitochondrial protein COX2 was found to be inversely related to that of its ribosome-bound mRNA.
Conclusions: Our results indicate that the association of mitochondrial mRNAs to ribosomes is largely determined by the total transcript level of a gene. However, at least for Cox2, we demonstrated that the level of ribosome-bound mRNA is not reflected by the amount of COX2 protein.
Keywords: Mitochondrial microarray; Mitochondrial ribosomes; Plant mitochondria; Transcription; Translation.
Figures




References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

