Mutations in HPV18 E1^E4 Impact Virus Capsid Assembly, Infectivity Competence, and Maturation
- PMID: 29257050
- PMCID: PMC5744159
- DOI: 10.3390/v9120385
Mutations in HPV18 E1^E4 Impact Virus Capsid Assembly, Infectivity Competence, and Maturation
Abstract
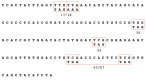
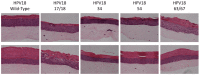
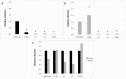
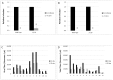
The most highly expressed protein during the productive phase of the human papillomavirus (HPV) life cycle is E1^E4. Its full role during infection remains to be established. HPV E1^E4 is expressed during both the early and late stages of the virus life cycle and contributes to viral genome amplification. In an attempt to further outline the functions of E1^E4, and determine whether it plays a role in viral capsid assembly and viral infectivity, we examined wild-type E1^E4 as well as four E1^E4 truncation mutants. Our study revealed that HPV18 genomes containing the shortest truncated form of E1^E4, the 17/18 mutant, produced viral titers that were similar to wild-type virus and significantly higher compared to virions containing the three longer E1^E4 mutants. Additionally, the infectivity of virus containing the shortest E1^E4 mutation was equivalent to wild-type and significantly higher than the other three mutants. In contrast, infectivity was completely abrogated for virus containing the longer E1^E4 mutants, regardless of virion maturity. Taken together, our results indicate for the first time that HPV18 E1^E4 impacts capsid assembly and viral infectivity as well as virus maturation.
Keywords: E1^E4; HPV18; Human Papillomavirus (HPV); infection; viral titer; virion maturation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
A cyclin-binding motif in human papillomavirus type 18 (HPV18) E1^E4 is necessary for association with CDK-cyclin complexes and G2/M cell cycle arrest of keratinocytes, but is not required for differentiation-dependent viral genome amplification or L1 capsid protein expression.Virology. 2011 Mar 30;412(1):196-210. doi: 10.1016/j.virol.2011.01.007. Epub 2011 Jan 31. Virology. 2011. PMID: 21276999 Free PMC article.
-
Uncovering the Role of the E1 Protein in Different Stages of Human Papillomavirus 18 Genome Replication.J Virol. 2020 Sep 29;94(20):e00674-20. doi: 10.1128/JVI.00674-20. Print 2020 Sep 29. J Virol. 2020. PMID: 32759324 Free PMC article.
-
HPV16 and 18 genome amplification show different E4-dependence, with 16E4 enhancing E1 nuclear accumulation and replicative efficiency via its cell cycle arrest and kinase activation functions.PLoS Pathog. 2017 Mar 17;13(3):e1006282. doi: 10.1371/journal.ppat.1006282. eCollection 2017 Mar. PLoS Pathog. 2017. PMID: 28306742 Free PMC article.
-
Study of infectious virus production from HPV18/16 capsid chimeras.Virology. 2010 Sep 30;405(2):289-99. doi: 10.1016/j.virol.2010.05.019. Epub 2010 Jul 3. Virology. 2010. PMID: 20598725 Free PMC article.
-
[Virological and carcinogenic aspects of HPV].Bull Acad Natl Med. 2007 Mar;191(3):611-23; discussion 623. Bull Acad Natl Med. 2007. PMID: 18072657 Review. French.
Cited by
-
The Role of Ataxia Telangiectasia Mutant and Rad3-Related DNA Damage Response in Pathogenesis of Human Papillomavirus.Pathogens. 2020 Jun 23;9(6):506. doi: 10.3390/pathogens9060506. Pathogens. 2020. PMID: 32585979 Free PMC article. Review.
-
Anti-Retroviral Protease Inhibitors Regulate Human Papillomavirus 16 Infection of Primary Oral and Cervical Epithelium.Cancers (Basel). 2020 Sep 18;12(9):2664. doi: 10.3390/cancers12092664. Cancers (Basel). 2020. PMID: 32961945 Free PMC article.
-
Manipulation of Epithelial Differentiation by HPV Oncoproteins.Viruses. 2019 Apr 22;11(4):369. doi: 10.3390/v11040369. Viruses. 2019. PMID: 31013597 Free PMC article. Review.
-
Assessing the reduction of viral infectivity in HPV16/18-positive women after one, two, and three doses of Gardasil-9 (RIFT): Study protocol.PLoS One. 2024 May 20;19(5):e0304080. doi: 10.1371/journal.pone.0304080. eCollection 2024. PLoS One. 2024. PMID: 38768231 Free PMC article.
-
Differential expression of human papillomavirus 16-, 18-, 52-, and 58-derived transcripts in cervical intraepithelial neoplasia.Virol J. 2020 Mar 6;17(1):32. doi: 10.1186/s12985-020-01306-0. Virol J. 2020. PMID: 32143682 Free PMC article.
References
-
- Syrjanen S., Lodi G., von Bultzingslowen I., Aliko A., Arduino P., Campisi G., Challacombe S., Ficarra G., Flaitz C., Zhou H.M., et al. Human papillomaviruses in oral carcinoma and oral potentially malignant disorders: A systematic review. Oral Dis. 2011;17(Suppl. 1):58–72. doi: 10.1111/j.1601-0825.2011.01792.x. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

