Twenty‑four signature genes predict the prognosis of oral squamous cell carcinoma with high accuracy and repeatability
- PMID: 29257303
- PMCID: PMC5783517
- DOI: 10.3892/mmr.2017.8256
Twenty‑four signature genes predict the prognosis of oral squamous cell carcinoma with high accuracy and repeatability
Abstract
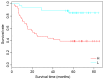
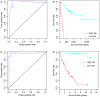
Oral squamous cell carcinoma (OSCC) is the sixth most common type cancer worldwide, with poor prognosis. The present study aimed to identify gene signatures that could classify OSCC and predict prognosis in different stages. A training data set (GSE41613) and two validation data sets (GSE42743 and GSE26549) were acquired from the online Gene Expression Omnibus database. In the training data set, patients were classified based on the tumor‑node‑metastasis staging system, and subsequently grouped into low stage (L) or high stage (H). Signature genes between L and H stages were selected by disparity index analysis, and classification was performed by the expression of these signature genes. The established classification was compared with the L and H classification, and fivefold cross validation was used to evaluate the stability. Enrichment analysis for the signature genes was implemented by the Database for Annotation, Visualization and Integration Discovery. Two validation data sets were used to determine the precise of classification. Survival analysis was conducted followed each classification using the package 'survival' in R software. A set of 24 signature genes was identified based on the classification model with the Fi value of 0.47, which was used to distinguish OSCC samples in two different stages. Overall survival of patients in the H stage was higher than those in the L stage. Signature genes were primarily enriched in 'ether lipid metabolism' pathway and biological processes such as 'positive regulation of adaptive immune response' and 'apoptotic cell clearance'. The results provided a novel 24‑gene set that may be used as biomarkers to predict OSCC prognosis with high accuracy, which may be used to determine an appropriate treatment program for patients with OSCC in addition to the traditional evaluation index.
Figures


References
-
- Wyss AB, Hashibe M, Lee YA, Chuang SC, Muscat J, Chen C, Schwartz SM, Smith E, Zhang ZF, Morgenstern H, et al. Smokeless Tobacco Use and the Risk of Head and Neck Cancer: Pooled Analysis of US Studies in the INHANCE Consortium. Am J Epidemiol. 2016 Oct 15; doi: 10.1093/aje/kww075. (Epub ahead of print) - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

