Multi-Omic Biogeography of the Gastrointestinal Microbiota of a Pre-Weaned Lamb
- PMID: 29258228
- PMCID: PMC5748571
- DOI: 10.3390/proteomes5040036
Multi-Omic Biogeography of the Gastrointestinal Microbiota of a Pre-Weaned Lamb
Abstract
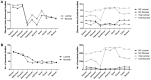
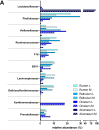
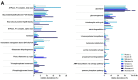
The digestive functions of the pre-weaned lamb gastrointestinal tracts (GITs) have been the subject of much research in recent years, but the microbial and host functions underlying these complex processes remain largely unknown. Here, we undertook a proof-of-principle metaproteogenomic investigation on luminal and mucosal samples collected from 10 GITs of a 30-day-old pre-weaned lamb. We demonstrate that the analysis of the diverse ecological niches along the GITs can reveal microbiota composition and metabolic functions, although low amounts of microbial proteins could be identified in the small intestinal and mucosal samples. Our data suggest that a 30-day lamb has already developed mature microbial functions in the forestomachs, while the effect of the milky diet appears to be more evident in the remaining GITs. We also report the distribution and the relative abundance of the host functions, active at the GIT level, with a special focus on those involved in digestive processes. In conclusion, this pilot study supports the suitability of a metaproteogenomic approach to the characterization of microbial and host functions of the lamb GITs, opening the way to further studies aimed at investigating the impact of early dietary interventions on the GIT microbiota of small ruminants.
Keywords: metaproteomics; microbial community; mucosa; ruminant; sheep.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

