Circadian clocks: from stem cells to tissue homeostasis and regeneration
- PMID: 29258993
- PMCID: PMC5757216
- DOI: 10.15252/embr.201745130
Circadian clocks: from stem cells to tissue homeostasis and regeneration
Abstract
The circadian clock is an evolutionarily conserved timekeeper that adapts body physiology to diurnal cycles of around 24 h by influencing a wide variety of processes such as sleep-to-wake transitions, feeding and fasting patterns, body temperature, and hormone regulation. The molecular clock machinery comprises a pathway that is driven by rhythmic docking of the transcription factors BMAL1 and CLOCK on clock-controlled output genes, which results in tissue-specific oscillatory gene expression programs. Genetic as well as environmental perturbation of the circadian clock has been implicated in various diseases ranging from sleep to metabolic disorders and cancer development. Here, we review the origination of circadian rhythms in stem cells and their function in differentiated cells and organs. We describe how clocks influence stem cell maintenance and organ physiology, as well as how rhythmicity affects lineage commitment, tissue regeneration, and aging.
Keywords: aging; circadian rhythms; clock; regeneration; stem cells.
© 2017 The Authors. Published under the terms of the CC BY NC ND 4.0 license.
Figures
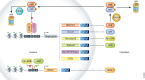
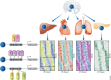

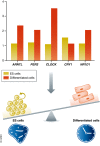
References
-
- Ebihara S, Tsuji K, Kondo K (1978) Strain differences of the mouse's free‐running circadian rhythm in continuous darkness. Physiol Behav 20: 795–799 - PubMed
-
- Balsalobre A, Damiola F, Schibler U (1998) A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93: 929–937 - PubMed
-
- Dierickx P, Du Pré B, Feyen DAM, Geijsen N, van Veen T, Doevendans PA, van Laake LW (2015) Circadian rhythms in stem cell biology and function In Stem Cells and Cardiac Regeneration, Madonna R, (ed.), pp 57–78. Cham: Springer International Publishing;
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

