Stepwise processing analyses of the single-turnover PCSK9 protease reveal its substrate sequence specificity and link clinical genotype to lipid phenotype
- PMID: 29259136
- PMCID: PMC5808750
- DOI: 10.1074/jbc.RA117.000754
Stepwise processing analyses of the single-turnover PCSK9 protease reveal its substrate sequence specificity and link clinical genotype to lipid phenotype
Erratum in
-
Stepwise processing analyses of the single-turnover PCSK9 protease reveal its substrate sequence specificity and link clinical genotype to lipid phenotype.J Biol Chem. 2018 May 4;293(18):6692. doi: 10.1074/jbc.AAC118.003335. J Biol Chem. 2018. PMID: 29728531 Free PMC article. No abstract available.
Abstract
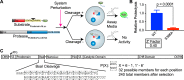
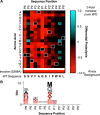
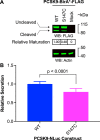
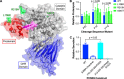
Proprotein convertase subtilisin/kexin type 9 (PCSK9) down-regulates the low-density lipoprotein (LDL) receptor, elevating LDL cholesterol and accelerating atherosclerotic heart disease, making it a promising cardiovascular drug target. To achieve its maximal effect on the LDL receptor, PCSK9 requires autoproteolysis. After cleavage, PCSK9 retains its prodomain in the active site as a self-inhibitor. Unlike other proprotein convertases, however, this retention is permanent, inhibiting any further protease activity for the remainder of its life cycle. Such inhibition has proven a major challenge toward a complete biochemical characterization of PCSK9's proteolytic function, which could inform therapeutic approaches against its hypercholesterolemic effects. To address this challenge, we employed a cell-based, high-throughput method using a luciferase readout to evaluate the single-turnover PCSK9 proteolytic event. We combined this method with saturation mutagenesis libraries to interrogate the sequence specificities of PCSK9 cleavage and proteolysis-independent secretion. Our results highlight several key differences in sequence identity between these two steps, complement known structural data, and suggest that PCSK9 self-proteolysis is the rate-limiting step of secretion. Additionally, we found that for missense SNPs within PCSK9, alterations in both proteolysis and secretion are common. Last, we show that some SNPs allosterically modulate PCSK9's substrate sequence specificity. Our findings indicate that PCSK9 proteolysis acts as a commonly perturbed but critical switch in controlling lipid homeostasis and provide a new hope for the development of small-molecule PCSK9 inhibitors.
Keywords: active site; atherosclerosis; high-throughput screening (HTS); low-density lipoprotein (LDL); proprotein convertase subtilisin/kexin type 9 (PCSK9); protein secretion; serine protease; single-nucleotide polymorphism (SNP); substrate specificity.
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures




References
-
- Zhang D.-W., Lagace T. A., Garuti R., Zhao Z., McDonald M., Horton J. D., Cohen J. C., and Hobbs H. H. (2007) Binding of proprotein convertase subtilisin/kexin type 9 to epidermal growth factor-like repeat A of low density lipoprotein receptor decreases receptor recycling and increases degradation. J. Biol. Chem. 282, 18602–18612 10.1074/jbc.M702027200 - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

